5CG0
 
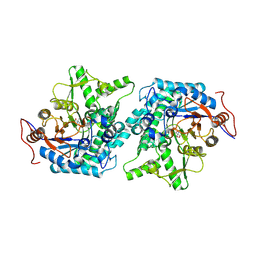 | | Crystal structure of Spodoptera frugiperda Beta-glycosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tamaki, F.K, Souza, D.P, Souza, V.P, Farah, S.C, Marana, S.R. | | Deposit date: | 2015-07-08 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Using the Amino Acid Network to Modulate the Hydrolytic Activity of beta-Glycosidases.
PLoS ONE, 11, 2016
|
|
2AKI
 
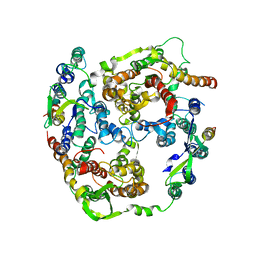 | | Normal mode-based flexible fitted coordinates of a translocating SecYEG protein-conducting channel into the cryo-EM map of a SecYEG-nascent chain-70S ribosome complex from E. coli | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, Protein-export membrane protein secG | | Authors: | Mitra, K, Schaffitzel, C, Shaikh, T, Tama, F, Jenni, S, Brooks III, C.L, Ban, N, Frank, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.9 Å) | | Cite: | Structure of the E. coli protein-conducting channel bound to a translating ribosome.
Nature, 438, 2005
|
|
2AKH
 
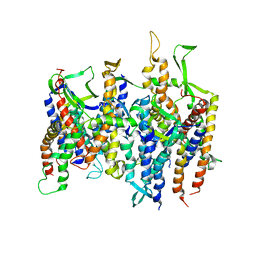 | | Normal mode-based flexible fitted coordinates of a non-translocating SecYEG protein-conducting channel into the cryo-EM map of a SecYEG-nascent chain-70S ribosome complex from E. coli | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, Protein-export membrane protein secG | | Authors: | Mitra, K.M, Schaffitzel, C, Shaikh, T, Tama, F, Jenni, S, Brooks III, C.L, Ban, N, Frank, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.9 Å) | | Cite: | Structure of the E. coli protein-conducting channel bound to a translating ribosome.
Nature, 438, 2005
|
|
4C3G
 
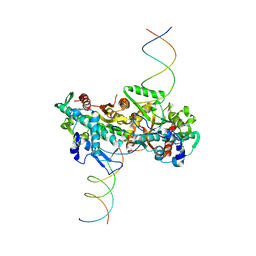 | | cryo-EM structure of activated and oligomeric restriction endonuclease SgrAI | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*GP*TP*GP*AP*AP*GP*AP*CP*CP *CP*AP*CP*GP*CP*AP*TP*CP)-3', 5'-D(*GP*AP*TP*GP*CP*GP*TP*GP*GP*GP*TP*CP*TP*TP *CP*AP*CP*AP)-3', SGRAIR RESTRICTION ENZYME | | Authors: | Lyumkis, D, Talley, H, Stewart, A, Shah, S, Park, C.K, Tama, F, Potter, C.S, Carragher, B, Horton, N.C. | | Deposit date: | 2013-08-23 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Allosteric Regulation of DNA Cleavage and Sequence-Specificity Through Run-on Oligomerization.
Structure, 21, 2013
|
|
6K8Q
 
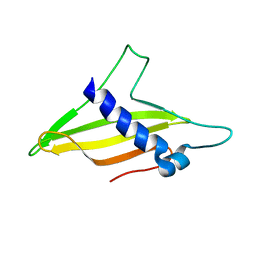 | | Solution structure of the intermembrane space domain of the mitochondrial import protein Tim21 from S. cerevisiae | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM21 | | Authors: | Bala, S, Shinya, S, Srivastava, A, Shimada, A, Kobayashi, N, Kojima, C, Tama, F, Miyashita, O, Kohda, D. | | Deposit date: | 2019-06-13 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
3J04
 
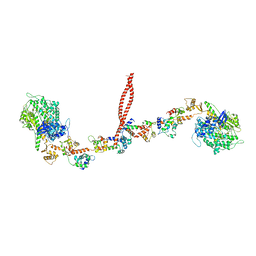 | | EM structure of the heavy meromyosin subfragment of Chick smooth muscle Myosin with regulatory light chain in phosphorylated state | | Descriptor: | Myosin light polypeptide 6, Myosin regulatory light chain 2, smooth muscle major isoform, ... | | Authors: | Baumann, B.A.J, Taylor, D, Huang, Z, Tama, F, Fagnant, P.M, Trybus, K, Taylor, K. | | Deposit date: | 2011-02-18 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Phosphorylated smooth muscle heavy meromyosin shows an open conformation linked to activation.
J.Mol.Biol., 415, 2012
|
|
1R4D
 
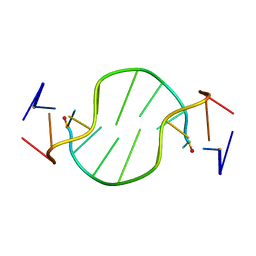 | | Solution structure of the chimeric L/D DNA oligonucleotide d(C8metGCGC(L)G(L)CGCG)2 | | Descriptor: | 5'-D(*CP*(8MG)P*CP*GP*(0DC)P*(0DG)P*CP*GP*CP*G)-3' | | Authors: | Cherrak, I, Mauffret, O, Santamaria, F, Rayner, B, Hocquet, A, Ghomi, M, Fermandjian, S. | | Deposit date: | 2003-10-06 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | L-nucleotides and 8-methylguanine of d(C1m8G2C3G4C5LG6LC7G8C9G10)2 act cooperatively to promote a left-handed helix under physiological salt conditions.
Nucleic Acids Res., 31, 2003
|
|
1FV8
 
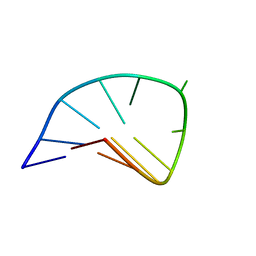 | | NMR STUDY OF AN HETEROCHIRAL HAIRPIN | | Descriptor: | 5'-D(*TP*AP*TP*CP*AP*(0DT)P*CP*GP*AP*TP*A)-3' | | Authors: | El Amri, C, Mauffret, O, Santamaria, F, Rayner, B, Fermandjian, S. | | Deposit date: | 2000-09-19 | | Release date: | 2000-10-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR study of a heterochiral DNA hairpin:impact of L-enantiomery in the loop.
J.Biomol.Struct.Dyn., 19, 2001
|
|
1FV7
 
 | | A TWO B-Z JUNCTION CONTAINING DNA RESOLVES INTO AN ALL RIGHT HANDED DOUBLE HELIX | | Descriptor: | 5'-D(*(5CM)P*GP*(5CM)P*GP*(0DC)P*(0DG)P*(5CM)P*GP*(5CM)P*G)-3' | | Authors: | Mauffret, O, El Amri, C, Santamaria, F, Tevanian, G, Rayner, B, Fermandjian, S. | | Deposit date: | 2000-09-19 | | Release date: | 2000-10-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A two B-Z junction containing DNA resolves into an all right-handed double-helix.
Nucleic Acids Res., 28, 2000
|
|
