3VRP
 
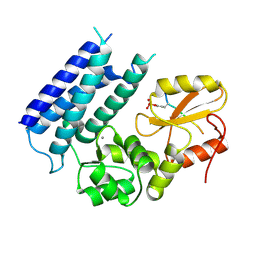 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c in complex with phospho-EGFR peptide | | Descriptor: | CALCIUM ION, Epidermal growth factor receptor, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3VRN
 
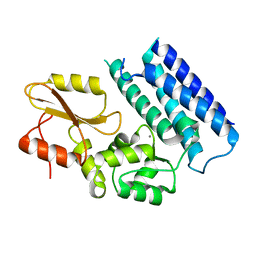 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c | | Descriptor: | CALCIUM ION, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c
J.Biochem., 152, 2012
|
|
3VRR
 
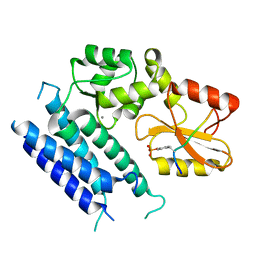 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c (PL mutant) in complex with phospho-EGFR peptide | | Descriptor: | CALCIUM ION, Epidermal growth factor receptor, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3VRO
 
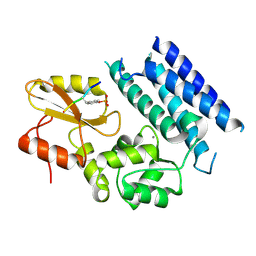 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c in complex with phospho-Src peptide | | Descriptor: | CALCIUM ION, Proto-oncogene tyrosine-protein kinase Src, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3VRQ
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c (PL mutant) | | Descriptor: | CALCIUM ION, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3WKV
 
 | | Voltage-gated proton channel: VSOP/Hv1 chimeric channel | | Descriptor: | Ion channel | | Authors: | Takeshita, K, Sakata, S, Yamashita, E, Fujiwara, Y, Kawanabe, A, Kurokawa, T, Okochi, Y, Matsuda, M, Narita, H, Okamura, Y, Nakagawa, A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.453 Å) | | Cite: | X-ray crystal structure of voltage-gated proton channel.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3AV4
 
 | | Crystal structure of mouse DNA methyltransferase 1 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AV6
 
 | | Crystal structure of mouse DNA methyltransferase 1 with AdoMet | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AV5
 
 | | Crystal structure of mouse DNA methyltransferase 1 with AdoHcy | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8JRD
 
 | | Chalcone synthase from Glycine max (L.) Merr (soybean) complexed with naringenin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Waki, T, Imaizumi, R, Nakata, S, Yanai, T, Takeshita, K, Sakai, N, Kataoka, K, Yamamoto, M, Nakayama, T, Yamashita, S. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural insights into catalytic promiscuity of chalcone synthase from Glycine max (L.) Merr.: Coenzyme A-induced alteration of product specificity.
Biochem.Biophys.Res.Commun., 718, 2024
|
|
7F19
 
 | | Structure of the G304E mutant of CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Takata, S.T, Kawano, T.K, Nakata, S.N, Takado, K.T, Imaizumi, R.I, Sakai, N.S, Takeshita, K.T, Yamashita, S.Y, Sakurai, T.S, Kataoka, K.K. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structure of the G304E mutant of CueO
To Be Published
|
|
7F0V
 
 | | Structure of the M305I mutant of CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Kawano, T.K, Takata, S.T, Sakai, N.S, Imaizumi, R.I, Nakata, S.N, Takeshita, K.T, Yamashita, S.Y, Sakurai, T.S, Kataoka, K.K. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | Structure of the M305I mutant of CueO
To Be Published
|
|
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
8X35
 
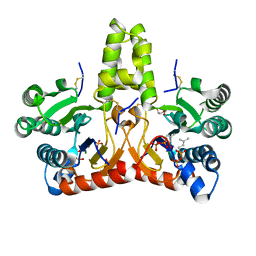 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP, IPP, and magnesium ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
8X37
 
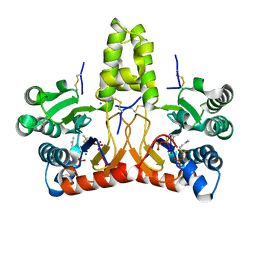 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, Neryl-diphosphate synthase 1 | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
7BUR
 
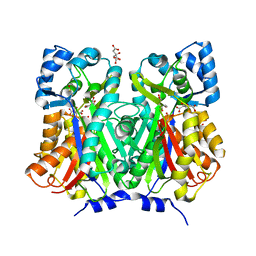 | | Chalcone synthase from Glycine max (L.) Merr (soybean) complexed with naringenin | | Descriptor: | CITRIC ACID, Chalcone synthase 1, NARINGENIN | | Authors: | Imaizumi, R, Mameda, R, Takeshita, K, Waki, T, Kubo, H, Sakai, N, Nakata, S, Takahashi, S, Kataoka, K, Yamamoto, M, Yamashita, S, Nakayama, T. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of chalcone synthase, a key enzyme for isoflavonoid biosynthesis in soybean.
Proteins, 2020
|
|
7BUS
 
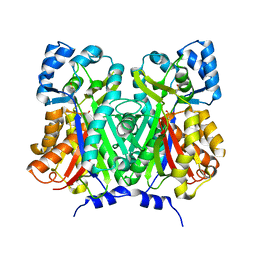 | | Chalcone synthase from Glycine max (L.) Merr (soybean) | | Descriptor: | Chalcone synthase | | Authors: | Imaizumi, R, Mameda, R, Takeshita, K, Waki, T, Kubo, H, Sakai, N, Nakata, S, Takahashi, S, Kataoka, K, Yamamoto, M, Yamashita, S, Nakayama, T. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of chalcone synthase, a key enzyme for isoflavonoid biosynthesis in soybean.
Proteins, 2020
|
|
8X36
 
 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP, IPP, and magnesium ion (form B) | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
3VMY
 
 | |
3VMX
 
 | | Crystal Structure of a parallel coiled-coil dimerization domain from the voltage-gated proton channel | | Descriptor: | Voltage-gated hydrogen channel 1 | | Authors: | Fujiwara, Y, Takeshita, K, Kobayashi, M, Okamura, Y, Nakagawa, A. | | Deposit date: | 2011-12-19 | | Release date: | 2012-05-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The cytoplasmic coiled-coil mediates cooperative gating temperature sensitivity in the voltage-gated H(+) channel Hv1
Nat Commun, 3, 2012
|
|
3VN0
 
 | |
3VMZ
 
 | |
2DQA
 
 | | Crystal Structure of Tapes japonica Lysozyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme, PLATINUM (II) ION, ... | | Authors: | Goto, T, Kakuta, Y, Abe, Y, Takeshita, K, Imoto, T, Ueda, T. | | Deposit date: | 2006-05-24 | | Release date: | 2007-06-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Tapes japonica Lysozyme with Substrate Analogue: STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND MANIFESTATION OF ITS CHITINASE ACTIVITY ACCOMPANIED BY QUATERNARY STRUCTURAL CHANGE
J.Biol.Chem., 282, 2007
|
|
7VPC
 
 | | Neryl diphosphate synthase from Solanum lycopersicum | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, Neryl-diphosphate synthase 1 | | Authors: | Imaizumi, R, Misawa, S, Takeshita, K, Sakai, N, Yamamoto, M, Kataoka, K, Nakayama, T, Takahashi, S, Yamashita, S. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-based engineering of a short-chain cis-prenyltransferase to biosynthesize nonnatural all-cis-polyisoprenoids: molecular mechanisms for primer substrate recognition and ultimate product chain-length determination.
Febs J., 289, 2022
|
|
5WY1
 
 | | Crystal structure of mouse DNA methyltransferase 1 (T1505A mutant) | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Kanada, K, Takeshita, K, Suetake, I, Tajima, S, Nakagawa, A. | | Deposit date: | 2017-01-10 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Conserved threonine 1505 in the catalytic domain stabilizes mouse DNA methyltransferase 1
J. Biochem., 162, 2017
|
|
