4W7H
 
 | | Crystal Structure of DEH Reductase A1-R Mutant | | Descriptor: | Carbonyl reductase | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
4W7I
 
 | | Crystal structure of DEH reductase A1-R' mutant | | Descriptor: | 4-deoxy-L-erythro-5-hexoseulose uronate reductase A1-R' | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
4Z9Y
 
 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Pectobacterium carotovorum | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase, SULFATE ION | | Authors: | Takase, R, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
4ZA2
 
 | | Crystal structure of Pectobacterium carotovorum 2-keto-3-deoxy-D-gluconate dehydrogenase complexed with NAD+ | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Takase, R, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
3AFM
 
 | | Crystal structure of aldose reductase A1-R responsible for alginate metabolism | | Descriptor: | Carbonyl reductase | | Authors: | Takase, R, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-03-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular identification of unsaturated uronate reductase prerequisite for alginate metabolism in Sphingomonas sp. A1
Biochim.Biophys.Acta, 1804, 2010
|
|
3AFN
 
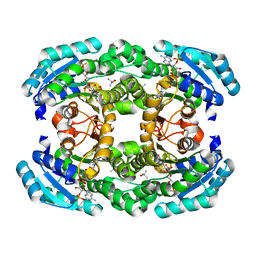 | | Crystal structure of aldose reductase A1-R complexed with NADP | | Descriptor: | Carbonyl reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TERTIARY-BUTYL ALCOHOL | | Authors: | Takase, R, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2010-03-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular identification of unsaturated uronate reductase prerequisite for alginate metabolism in Sphingomonas sp. A1
Biochim.Biophys.Acta, 1804, 2010
|
|
4TKL
 
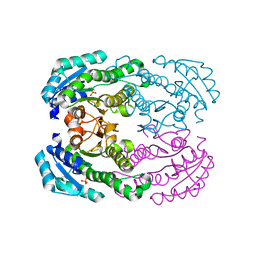 | | Crystal structure of NADH-dependent reductase A1-R' responsible for alginate metabolism | | Descriptor: | NADH-dependent reductase for 4-deoxy-L-erythro-5-hexoseulose uronate, PHOSPHATE ION | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
4TKM
 
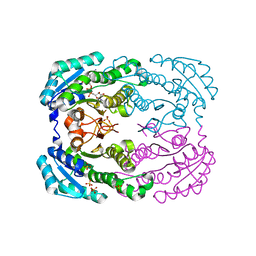 | | Crystal structure of NADH-dependent reductase A1-R' complexed with NAD | | Descriptor: | NADH-dependent reductase for 4-deoxy-L-erythro-5-hexoseulose uronate, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
6JBN
 
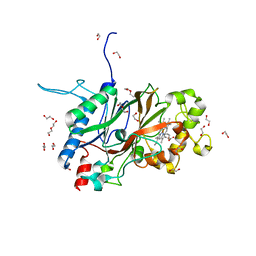 | | Crystal structure of Sphingomonas sp. A1 peroxidase EfeB responsible for import of iron | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, OXYGEN MOLECULE, ... | | Authors: | Okumura, K, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2019-01-26 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rare metal binding by a cell-surface component of bacterial EfeUOB iron importer
To Be Published
|
|
6JBO
 
 | | Crystal structure of EfeO-like protein Algp7 containing samarium ion | | Descriptor: | 1,2-ETHANEDIOL, Alginate-binding protein, CITRIC ACID | | Authors: | Okumura, K, Takase, R, Maruyama, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2019-01-26 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Rare metal binding by a cell-surface component of bacterial EfeUOB iron importer
To Be Published
|
|
4Z9X
 
 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Streptococcus pyogenes | | Descriptor: | Gluconate 5-dehydrogenase | | Authors: | Maruyama, Y, Takase, R, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
4U8F
 
 | | Crystal structure of 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase complexed with a tartrate | | Descriptor: | L(+)-TARTARIC ACID, Putative uncharacterized protein gbs1892 | | Authors: | Maruyama, Y, Oiki, S, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Metabolic Fate of Unsaturated Glucuronic/Iduronic Acids from Glycosaminoglycans: MOLECULAR IDENTIFICATION AND STRUCTURE DETERMINATION OF STREPTOCOCCAL ISOMERASE AND DEHYDROGENASE.
J.Biol.Chem., 290, 2015
|
|
4U8G
 
 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Streptococcus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1891 | | Authors: | Maruyama, Y, Oiki, S, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Metabolic Fate of Unsaturated Glucuronic/Iduronic Acids from Glycosaminoglycans: MOLECULAR IDENTIFICATION AND STRUCTURE DETERMINATION OF STREPTOCOCCAL ISOMERASE AND DEHYDROGENASE.
J.Biol.Chem., 290, 2015
|
|
4U8E
 
 | | Crystal structure of 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase from Streptococcus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1892 | | Authors: | Maruyama, y, Oiki, S, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-03 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolic Fate of Unsaturated Glucuronic/Iduronic Acids from Glycosaminoglycans: MOLECULAR IDENTIFICATION AND STRUCTURE DETERMINATION OF STREPTOCOCCAL ISOMERASE AND DEHYDROGENASE
J.Biol.Chem., 290, 2015
|
|
7E4S
 
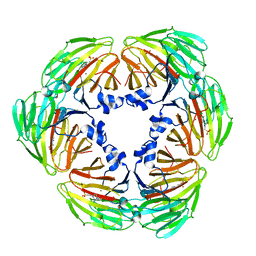 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-dehydro-4-deoxy-D-glucuronate isomerase, ZINC ION | | Authors: | Yamamoto, Y, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-02-15 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structures of Lacticaseibacillus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI in complex with substrate analogs
J.Appl.Glyosci., 2023
|
|
7YE3
 
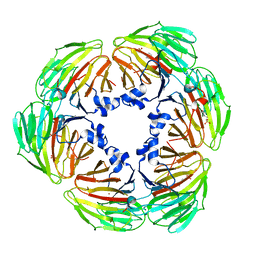 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
7YRS
 
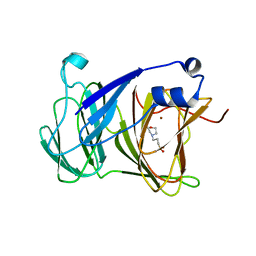 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MOPS | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
7VEW
 
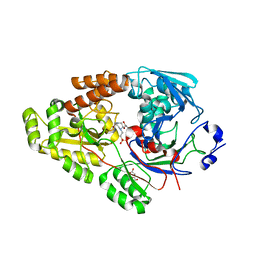 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with unsaturated trigalacturonic acid | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEQ
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in an open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VET
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a closed conformation | | Descriptor: | SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEV
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VER
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a full open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEU
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with galacturonic acid | | Descriptor: | GLYCEROL, SPH1118, alpha-D-galactopyranuronic acid | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7WGU
 
 | | Crystal structure of metal-binding protein EfeO from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Iron uptake system protein EfeO, ... | | Authors: | Nakatsuji, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-12-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of EfeB and EfeO in a bacterial siderophore-independent iron transport system
Biochem.Biophys.Res.Commun., 594, 2022
|
|
7VGK
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI | | Descriptor: | 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase | | Authors: | Iwase, H, Oiki, S, Mikami, B, Takase, R, Hashimoto, W. | | Deposit date: | 2021-09-16 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of Lacticaseibacillus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI in complex with substrate analogs
J.Appl.Glyosci., 2023
|
|
