1IYY
 
 | | NMR STRUCTURE OF Gln25-RIBONUCLEASE T1, 24 STRUCTURES | | Descriptor: | RIBONUCLEASE T1 | | Authors: | Hatano, K, Kojima, M, Suzuki, E, Tanokura, M, Takahashi, K. | | Deposit date: | 2002-09-12 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Determination of the NMR structure of Gln25-ribonuclease T1.
Biol. Chem., 384, 2003
|
|
7ZPZ
 
 | | Crystal structure of Pizza6-TSR-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-TSR-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
7ZQ2
 
 | | Crystal structure of Pizza6-RSH-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-RSH-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
7ZQG
 
 | | Crystal structure of Pizza6-KSH-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-KSH-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
7ZPW
 
 | | Crystal structure of Pizza6-TSK-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-TSK-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface.
To be published
|
|
7ZPH
 
 | | Crystal structure of Pizza6-TNK-RSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-TNK-RSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-27 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
7ZCJ
 
 | | Crystal structure of Pizza6-TNH-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-TNH-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-03-28 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
1DET
 
 | | RIBONUCLEASE T1 CARBOXYMETHYLATED AT GLU 58 IN COMPLEX WITH 2'GMP | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Ishikawa, K, Suzuki, E, Tanokura, M, Takahashi, K. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ribonuclease T1 carboxymethylated at Glu58 in complex with 2'-GMP.
Biochemistry, 35, 1996
|
|
1TFK
 
 | | Ribonuclease from Escherichia coli complexed with its inhibtor protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Colicin D, Colicin D immunity protein | | Authors: | Yajima, S, Nakanishi, K, Takahashi, K, Ogawa, T, Kezuka, Y, Hidaka, M, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2004-05-27 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Relation between tRNase activity and the structure of colicin D according to X-ray crystallography
Biochem.Biophys.Res.Commun., 322, 2004
|
|
1TFO
 
 | | Ribonuclease from Escherichia coli complexed with its inhibitor protein | | Descriptor: | Colicin D, Colicin D immunity protein | | Authors: | Yajima, S, Nakanishi, K, Takahashi, K, Ogawa, T, Kezuka, Y, Hidaka, M, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2004-05-27 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relation between tRNase activity and the structure of colicin D according to X-ray crystallography
Biochem.Biophys.Res.Commun., 322, 2004
|
|
1TAB
 
 | | STRUCTURE OF THE TRYPSIN-BINDING DOMAIN OF BOWMAN-BIRK TYPE PROTEASE INHIBITOR AND ITS INTERACTION WITH TRYPSIN | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR, TRYPSIN | | Authors: | Tsunogae, Y, Tanaka, I, Yamane, T, Kikkawa, J.-I, Ashida, T, Ishikawa, C, Watanabe, K, Nakamura, S, Takahashi, K. | | Deposit date: | 1990-10-15 | | Release date: | 1992-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the trypsin-binding domain of Bowman-Birk type protease inhibitor and its interaction with trypsin.
J.Biochem.(Tokyo), 100, 1986
|
|
1IPJ
 
 | | CRYSTAL STRUCTURES OF RECOMBINANT AND NATIVE SOYBEAN BETA-CONGLYCININ BETA HOMOTRIMERS COMPLEXES WITH N-ACETYL-D-GLUCOSAMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-CONGLYCININ, BETA CHAIN | | Authors: | Maruyama, N, Adachi, M, Takahashi, K, Yagasaki, K, Kohno, M, Takenaka, Y, Okuda, E, Nakagawa, S, Mikami, B, Utsumi, S. | | Deposit date: | 2001-05-16 | | Release date: | 2002-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of recombinant and native soybean beta-conglycinin beta homotrimers.
Eur.J.Biochem., 268, 2001
|
|
1IPK
 
 | | CRYSTAL STRUCTURES OF RECOMBINANT AND NATIVE SOYBEAN BETA-CONGLYCININ BETA HOMOTRIMERS | | Descriptor: | BETA-CONGLYCININ, BETA CHAIN | | Authors: | Maruyama, N, Adachi, M, Takahashi, K, Yagasaki, K, Kohno, M, Takenaka, Y, Okuda, E, Nakagawa, S, Mikami, B, Utsumi, S. | | Deposit date: | 2001-05-16 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of recombinant and native soybean beta-conglycinin beta homotrimers.
Eur.J.Biochem., 268, 2001
|
|
7WAB
 
 | | Crystal structure of the prolyl endoprotease, PEP, from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPASS (Complex proteins associated with Set1p) component shg1 family protein, ... | | Authors: | Miyazono, K, Kubota, K, Takahashi, K, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and substrate recognition mechanism of the prolyl endoprotease PEP from Aspergillus niger.
Biochem.Biophys.Res.Commun., 591, 2022
|
|
2LHI
 
 | | Solution structure of Ca2+/CNA1 peptide-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin,Serine/threonine-protein phosphatase 2B catalytic subunit A1 | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
2LHH
 
 | | Solution structure of Ca2+-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
7VUS
 
 | | Crystal structure of AlleyCat9 with 5-nitro-benzotriazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-nitro-1H-benzotriazole, AlleyCat, ... | | Authors: | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7VUR
 
 | | Crystal structure of AlleyCat9 with calcium but no inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AlleyCat, CALCIUM ION | | Authors: | Margheritis, E, Takahashi, K, Korendovych, I.V, Tame, J.R.H. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7VUU
 
 | | Crystal structure of AlleyCat10 with inhibitor | | Descriptor: | 5-nitro-1H-benzotriazole, AlleyCat, CALCIUM ION | | Authors: | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7VUT
 
 | | Crystal structure of AlleyCat10 | | Descriptor: | AlleyCat10, CALCIUM ION | | Authors: | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
3TRS
 
 | | The crystal structure of aspergilloglutamic peptidase from Aspergillus niger | | Descriptor: | Aspergillopepsin-2 heavy chain, Aspergillopepsin-2 light chain, DIMETHYL SULFOXIDE | | Authors: | Sasaki, H, Kubota, K, Lee, W.C, Ohtsuka, J, Kojima, M, Takahashi, K, Tanokura, M. | | Deposit date: | 2011-09-10 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of an intermediate dimer of aspergilloglutamic peptidase that mimics the enzyme-activation product complex produced upon autoproteolysis.
J.Biochem., 152, 2012
|
|
2D1A
 
 | | Solution RNA structure model of the HIV-1 dimerization initiation site in the extended-duplex dimer | | Descriptor: | RNA | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2D17
 
 | | Solution RNA structure of stem-bulge-stem region of the HIV-1 dimerization initiation site | | Descriptor: | 5'-R(*CP*GP*GP*CP*AP*AP*GP*AP*GP*GP*CP*GP*AP*CP*CP*C)-3', 5'-R(*GP*GP*GP*UP*CP*GP*GP*CP*UP*UP*GP*CP*UP*G)-3' | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2D1B
 
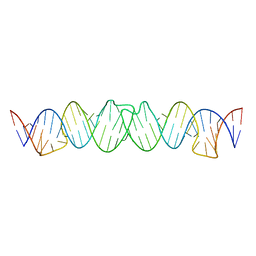 | | Solution RNA structure model of the HIV-1 dimerization initiation site in the kissing-loop dimer | | Descriptor: | RNA | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2D18
 
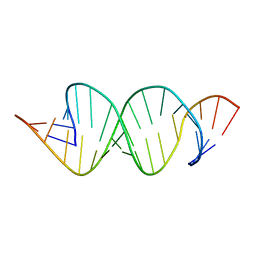 | | Solution RNA structure of loop region of the HIV-1 dimerization initiation site in the extended-duplex dimer | | Descriptor: | 5'-R(*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*GP*GP*C)-3' | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
