5T6D
 
 | | 2.10 A resolution structure of Norovirus 3CL protease in complex with the dipeptidyl inhibitor 7l (hexagonal form) | | Descriptor: | 3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[3-(4-methoxyphenoxy)propyl]sulfonyl}-L- alaninamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Damalanka, V.C, Weerawarna, P.M, Doyle, S.T, Alsoudi, A.F, Dissanayake, D.M.P, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors.
Eur J Med Chem, 126, 2016
|
|
5T6G
 
 | | 2.45 A resolution structure of Norovirus 3CL protease in complex with the dipeptidyl inhibitor 7m (hexagonal form) | | Descriptor: | 3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-(octylsulfonyl)-L-alaninamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Damalanka, V.C, Weerawarna, P.M, Doyle, S.T, Alsoudi, A.F, Dissanayake, D.M.P, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors.
Eur J Med Chem, 126, 2016
|
|
5TG1
 
 | | 1.40 A resolution structure of Norovirus 3CL protease in complex with the a m-chlorophenyl substituted macrocyclic inhibitor (17-mer) | | Descriptor: | (4S,7S,17S)-17-(3-chlorophenyl)-7-(hydroxymethyl)-4-(2-methylpropyl)-1-oxa-3,6,11-triazacycloheptadecane-2,5,10-trione, 3C-LIKE PROTEASE, CHLORIDE ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Kankanamalage, A.C.G, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis, and evaluation of a novel series of macrocyclic inhibitors of norovirus 3CL protease.
Eur J Med Chem, 127, 2016
|
|
5TG2
 
 | | 1.75 A resolution structure of Norovirus 3CL protease in complex with the a n-pentyl substituted macrocyclic inhibitor (17-mer) | | Descriptor: | (4S,7S,17R)-7-(hydroxymethyl)-4-(2-methylpropyl)-17-pentyl-1-oxa-3,6,11-triazacycloheptadecane-2,5,10-trione, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Kankanamalage, A.C.G, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis, and evaluation of a novel series of macrocyclic inhibitors of norovirus 3CL protease.
Eur J Med Chem, 127, 2016
|
|
8G1W
 
 | | Crystal Structure Matriptase (C731S) in Complex with Inhibitor VD4162B | | Descriptor: | CHLORIDE ION, Cyclic peptide inhibitor (ACE)Y(DTR)(NLE)(KCM), Cyclic peptide inhibitor (ACE)Y(DTR)(NLE)(THZ), ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Janetka, J.W. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mechanism-Based Macrocyclic Inhibitors of Serine Proteases.
J.Med.Chem., 67, 2024
|
|
3MOM
 
 | | Structure of holo HasAp H32A mutant complexed with imidazole from Pseudomonas aeruginosa to 2.25A Resolution | | Descriptor: | Heme acquisition protein HasAp, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Jepkorir, G, Rodriguez, J.C, Rui, H, Im, W, Alontaga, A.Y, Yukl, E, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2010-04-22 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural, NMR Spectroscopic, and Computational Investigation of Hemin Loading in the Hemophore HasAp from Pseudomonas aeruginosa.
J.Am.Chem.Soc., 132, 2010
|
|
3MOL
 
 | | Structure of dimeric holo HasAp H32A Mutant from Pseudomonas aeruginosa to 1.20A Resolution | | Descriptor: | Heme acquisition protein HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Jepkorir, G, Rodriguez, J.C, Rui, H, Im, W, Alontaga, A.Y, Yukl, E, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2010-04-22 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural, NMR Spectroscopic, and Computational Investigation of Hemin Loading in the Hemophore HasAp from Pseudomonas aeruginosa.
J.Am.Chem.Soc., 132, 2010
|
|
3MOK
 
 | | Structure of Apo HasAp from Pseudomonas aeruginosa to 1.55A Resolution | | Descriptor: | Heme acquisition protein HasAp, PHOSPHATE ION, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Jepkorir, G, Rodriguez, J.C, Rui, H, Im, W, Alontaga, A.Y, Yukl, E, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2010-04-22 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, NMR Spectroscopic, and Computational Investigation of Hemin Loading in the Hemophore HasAp from Pseudomonas aeruginosa.
J.Am.Chem.Soc., 132, 2010
|
|
1NBK
 
 | | The structure of RNA aptamer for HIV Tat complexed with two argininamide molecules | | Descriptor: | 2-AMINO-5-GUANIDINO-PENTANOIC ACID, RNA aptamer | | Authors: | Matsugami, A, Kobayashi, S, Ouhashi, K, Uesugi, S, Yamamoto, R, Taira, K, Nishikawa, S, Kumar, P.K.R, Katahira, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2024-09-18 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Highly Efficient Trapping of the HIV Tat Protein by an RNA Aptamer
Structure, 11, 2003
|
|
8VIT
 
 | | Crystal structure of the N-terminal domain of fatty acid kinase A (FakA) from Staphylococcus aureus (Mg and ADP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FATTY ACID KINASE A, MAGNESIUM ION, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Bose, J.L. | | Deposit date: | 2024-01-05 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular insights into the structure and function of the Staphylococcus aureus fatty acid kinase
J.Biol.Chem., 2024
|
|
8VIR
 
 | |
8VIP
 
 | | Crystal structure of the N-terminal domain of fatty acid kinase A (FakA) from Staphylococcus aureus (Mn and AMP-PNP) | | Descriptor: | FATTY ACID KINASE A, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Bose, J.L. | | Deposit date: | 2024-01-05 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Molecular insights into the structure and function of the Staphylococcus aureus fatty acid kinase
J.Biol.Chem., 2024
|
|
8VIQ
 
 | | Crystal structure of the N-terminal domain of fatty acid kinase A (FakA) from Staphylococcus aureus (Mg and AMP-PNP) | | Descriptor: | FATTY ACID KINASE A, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Bose, J.L. | | Deposit date: | 2024-01-05 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular insights into the structure and function of the Staphylococcus aureus fatty acid kinase
J.Biol.Chem., 2024
|
|
2HA8
 
 | | Methyltransferase Domain of Human TAR (HIV-1) RNA binding protein 1 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, TAR (HIV-1) RNA loop binding protein | | Authors: | Min, J, Wu, H, Zeng, H, Loppnau, P, Battaile, K, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Methyltransferase Domain of Human TAR (HIV-1) RNA binding protein 1 in complex with S-adenosyl-L-homocystein.
To be Published
|
|
3SZE
 
 | | Crystal structure of the passenger domain of the E. coli autotransporter EspP | | Descriptor: | Serine protease espP | | Authors: | Khan, S, Mian, H.S, Sandercock, L.E, Battaile, K.P, Lam, R, Chirgadze, N.Y, Pai, E.F. | | Deposit date: | 2011-07-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Passenger Domain of the Escherichia coli Autotransporter EspP.
J.Mol.Biol., 413, 2011
|
|
2D24
 
 | | Crystal structure of ES complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D22
 
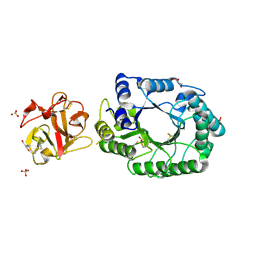 | | Crystal structure of covalent glycosyl-enzyme intermediate of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2Z5D
 
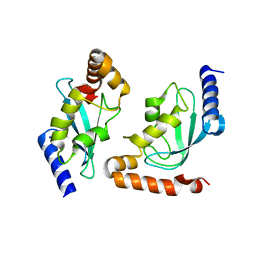 | | Human ubiquitin-conjugating enzyme E2 H | | Descriptor: | SODIUM ION, Ubiquitin-conjugating enzyme E2 H | | Authors: | Bochkarev, A, Cui, H, Walker, J.R, Newman, E.M, Mackenzie, F, Battaile, K.P, Sundstrom, M, Arrowsmith, C, Edwards, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
MOL.CELL PROTEOMICS, 11, 2012
|
|
2Q5R
 
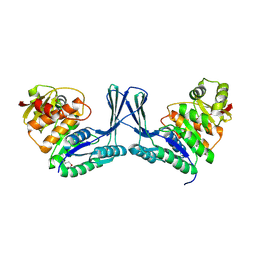 | | Structure of apo Staphylococcus aureus D-tagatose-6-phosphate kinase | | Descriptor: | Tagatose-6-phosphate kinase | | Authors: | McGrath, T.E, Soloveychik, M, Romanov, V, Thambipillai, D, Dharamsi, A, Virag, C, Domagala, M, Pai, E.F, Edwards, A.M, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2007-06-01 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of apo Staphylococcus aureus D-tagatose-6-phosphate kinase
TO BE PUBLISHED
|
|
3IS7
 
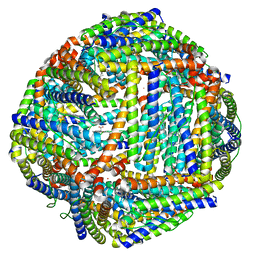 | | Structure of mineralized Bfrb from Pseudomonas aeruginosa to 2.1A Resolution | | Descriptor: | Bacterioferritin, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Weeratunga, S.K, Battaile, K.P, Rivera, M. | | Deposit date: | 2009-08-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Studies of Bacterioferritin B from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center
Biochemistry, 49, 2010
|
|
3ISE
 
 | | Structure of mineralized Bfrb (double soak) from Pseudomonas aeruginosa to 2.8A Resolution | | Descriptor: | Bacterioferritin, FE (III) ION, POTASSIUM ION, ... | | Authors: | Lovell, S, Weeratunga, S.K, Battaile, K.P, Rivera, M. | | Deposit date: | 2009-08-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of Bacterioferritin B from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center
Biochemistry, 49, 2010
|
|
3IS8
 
 | | Structure of mineralized Bfrb soaked with FeSO4 from Pseudomonas aeruginosa to 2.25A Resolution | | Descriptor: | Bacterioferritin, FE (II) ION, POTASSIUM ION, ... | | Authors: | Lovell, S, Weeratunga, S.K, Battaile, K.P, Rivera, M. | | Deposit date: | 2009-08-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Studies of Bacterioferritin B from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center
Biochemistry, 49, 2010
|
|
3ISF
 
 | | Structure of non-mineralized Bfrb (as-isolated) from Pseudomonas aeruginosa to 2.07A Resolution | | Descriptor: | Bacterioferritin, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Weeratunga, S.K, Battaile, K.P, Rivera, M. | | Deposit date: | 2009-08-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Studies of Bacterioferritin B from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center
Biochemistry, 49, 2010
|
|
3GPH
 
 | |
3IER
 
 | | Firefly luciferase apo structure (P41 form) with PEG 400 bound | | Descriptor: | Luciferin 4-monooxygenase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Auld, D.S, Thorne, N, Lea, W.A, Maloney, D.J, Shen, M, Raj, G, Thomas, C.J, Simeonov, A, Hanzlik, R.P, Inglese, J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
