7XJE
 
 | | Crystal structure of bacteriorhodopsin in the K state refined against the extrapolated dataset | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Taguchi, S, Niwa, S, Takeda, K. | | Deposit date: | 2022-04-16 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Detailed analysis of distorted retinal and its interaction with surrounding residues in the K intermediate of bacteriorhodopsin
Commun Biol, 6, 2023
|
|
7XJC
 
 | | Crystal structure of bacteriorhodopsin in the ground and K states after green laser irradiation | | Descriptor: | 2,10,23-TRIMETHYL-TETRACOSANE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Taguchi, S, Niwa, S, Takeda, K. | | Deposit date: | 2022-04-16 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Detailed analysis of distorted retinal and its interaction with surrounding residues in the K intermediate of bacteriorhodopsin
Commun Biol, 6, 2023
|
|
7XJD
 
 | | Crystal structure of bacteriorhodopsin in the ground state by red laser irradiation | | Descriptor: | 2,10,23-TRIMETHYL-TETRACOSANE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Taguchi, S, Niwa, S, Takeda, K. | | Deposit date: | 2022-04-16 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Detailed analysis of distorted retinal and its interaction with surrounding residues in the K intermediate of bacteriorhodopsin.
Commun Biol, 6, 2023
|
|
7X4N
 
 | | Crystal Structure of C. elegans kinesin-4 KLP-12 complexed with tubulin and DARPin | | Descriptor: | DARPin, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Taguchi, S, Imasaki, T, Saijo-Hamano, Y, Sakai, N, Nitta, R. | | Deposit date: | 2022-03-03 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural model of microtubule dynamics inhibition by kinesin-4 from the crystal structure of KLP-12 -tubulin complex.
Elife, 11, 2022
|
|
2RNE
 
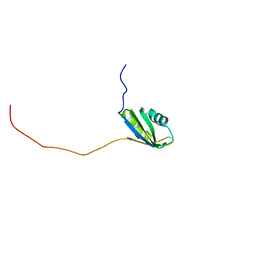 | | Solution structure of the second RNA recognition motif (RRM) of TIA-1 | | Descriptor: | Tia1 protein | | Authors: | Takahashi, M, Kuwasako, K, Abe, C, Tsuda, K, Inoue, M, Terada, T, Shirouzu, M, Kobayashi, N, Kigawa, T, Taguchi, S, Guntert, P, Hayashizaki, Y, Tanaka, A, Muto, Y, Yokoyama, S. | | Deposit date: | 2007-12-19 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RNA recognition motif (RRM) domain of murine T cell intracellular antigen-1 (TIA-1) and its RNA recognition mode
Biochemistry, 47, 2008
|
|
6AA8
 
 | |
6ACQ
 
 | |
