3WNU
 
 | | The crystal structure of catalase-peroxidase, KatG, from Synechococcus PCC7942 | | Descriptor: | Catalase-peroxidase, HEME B/C, SODIUM ION | | Authors: | Tada, T, Wada, K, Kamachi, S. | | Deposit date: | 2013-12-17 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 angstrom resolution structure of the catalase-peroxidase KatG from Synechococcus elongatus PCC7942.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3X16
 
 | |
4PAE
 
 | | Crystal structure of catalase-peroxidase (KatG) W78F mutant from Synechococcus elongatus PCC7942 | | Descriptor: | Catalase-peroxidase, HEME B/C, SODIUM ION, ... | | Authors: | Kamachi, S, Wada, K, Tada, T. | | Deposit date: | 2014-04-08 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Crystal structure of the catalase-peroxidase KatG W78F mutant from Synechococcus elongatus PCC7942 in complex with the antitubercular pro-drug isoniazid.
Febs Lett., 589, 2015
|
|
1QR3
 
 | | Structure of porcine pancreatic elastase in complex with FR901277, a novel macrocyclic inhibitor of elastases at 1.6 angstrom resolution | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, FR901277 Inhibitor, ... | | Authors: | Nakanishi, I, Kinoshita, T, Sato, A, Tada, T. | | Deposit date: | 1999-06-18 | | Release date: | 2000-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of porcine pancreatic elastase complexed with FR901277, a novel macrocyclic inhibitor of elastases, at 1.6 A resolution.
Biopolymers, 53, 2000
|
|
3VN9
 
 | | Rifined Crystal structure of non-phosphorylated MAP2K6 in a putative auto-inhibition state | | Descriptor: | 9-{5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]-beta-L-ribofuranosyl}-9H-purin-6-amine, Dual specificity mitogen-activated protein kinase kinase 6, MAGNESIUM ION | | Authors: | Kinoshita, T, Matsuzaka, H, Nakai, R, Kirii, Y, Yokota, K, Tada, T, Matsumoto, T. | | Deposit date: | 2012-01-05 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of non-phosphorylated MAP2K6 in a putative auto-inhibition state
J.Biochem., 151, 2012
|
|
3VUT
 
 | | Crystal structures of non-phosphorylated MAP2K4 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 4 | | Authors: | Matsumoto, T, Kinoshita, T, Kirii, Y, Tada, T, Yamano, A. | | Deposit date: | 2012-07-05 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal and solution structures disclose a putative transient state of mitogen-activated protein kinase kinase 4
Biochem.Biophys.Res.Commun., 425, 2012
|
|
6A8H
 
 | | Crystal structure of endo-arabinanase ABN-TS D27A mutant in complex with arabinotriose | | Descriptor: | CALCIUM ION, MAGNESIUM ION, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, ... | | Authors: | Yamaguchi, A, Tada, T. | | Deposit date: | 2018-07-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of endo-1,5-alpha-L-arabinanase mutants from Bacillus thermodenitrificans TS-3 in complex with arabino-oligosaccharides.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3HGN
 
 | | Structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180 determined by neutron crystallography | | Descriptor: | 4-[[(2S)-3-methyl-1-oxo-1-[(2S)-2-[[(3S)-1,1,1-trifluoro-4-methyl-2-oxo-pentan-3-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamoyl]benzoic acid, CALCIUM ION, Elastase-1, ... | | Authors: | Tamada, T, Kinoshita, T, Kuroki, R, Tada, T. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (1.65 Å), X-RAY DIFFRACTION | | Cite: | Combined High-Resolution Neutron and X-ray Analysis of Inhibited Elastase Confirms the Active-Site Oxyanion Hole but Rules against a Low-Barrier Hydrogen Bond
J.Am.Chem.Soc., 131, 2009
|
|
1VBL
 
 | | Structure of the thermostable pectate lyase PL 47 | | Descriptor: | CALCIUM ION, pectate lyase 47 | | Authors: | Nakaniwa, T, Tada, T, Yamaguchi, A, Kitatani, T, Takao, M, Sakai, T, Nishimura, K. | | Deposit date: | 2004-02-27 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Basis for Thermostability of Pectate Lyase from Bacillus sp. TS 47
To be Published
|
|
3HGP
 
 | | Structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180 determined by high resolution crystallography | | Descriptor: | 4-[[(2S)-3-methyl-1-oxo-1-[(2S)-2-[[(3S)-1,1,1-trifluoro-4-methyl-2-oxo-pentan-3-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamoyl]benzoic acid, CALCIUM ION, Elastase-1, ... | | Authors: | Tamada, T, Kinoshita, T, Kuroki, R, Tada, T. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Combined High-Resolution Neutron and X-ray Analysis of Inhibited Elastase Confirms the Active-Site Oxyanion Hole but Rules against a Low-Barrier Hydrogen Bond
J.Am.Chem.Soc., 131, 2009
|
|
3E3B
 
 | |
1UB2
 
 | |
1WL7
 
 | | Structure of the thermostable arabinanase | | Descriptor: | CALCIUM ION, arabinanase-TS | | Authors: | Yamaguchi, A, Tada, T, Nakaniwa, T, Kitatani, T. | | Deposit date: | 2004-06-21 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for thermostability of endo-1,5-alpha-L-arabinanase from Bacillus thermodenitrificans TS-3.
J.Biochem.(Tokyo), 137, 2005
|
|
6IGU
 
 | | Crystal structure of the hydrolytic antibody Fab 9C10 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IMMUNOGLOBULIN 9C10 H CHAIN, IMMUNOGLOBULIN 9C10 L CHAIN, ... | | Authors: | Yamaguchi, A, Tada, T, Tsuchiya, Y, Tsumuraya, T, Fujii, I. | | Deposit date: | 2018-09-26 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of the complex of the hydrolytic antibody Fab 9C10 and a transition-state analog
To Be Published
|
|
5XQJ
 
 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose substituted with galactose | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-[beta-D-galactopyranose-(1-4)]alpha-L-rhamnopyranose, 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, CALCIUM ION, ... | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
5XQG
 
 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, CALCIUM ION, Pcrglx protein | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
3VOC
 
 | | Crystal structure of the catalytic domain of beta-amylase from paenibacillus polymyxa | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta/alpha-amylase, ... | | Authors: | Nishimura, S, Fujioka, T, Nakaniwa, T, Tada, T. | | Deposit date: | 2012-01-21 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis by X-ray crystallography and small-angle scattering of the multi-domain beta-amylase from Paenibacillus polymyxa
To be Published
|
|
5XQO
 
 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with tetrameric substrate | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose-(1-4)-alpha-D-galactopyranuronic acid-(1-2)-alpha-L-rhamnopyranose, 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-alpha-L-rhamnopyranose-(1-4)-alpha-D-galactopyranuronic acid-(1-2)-alpha-L-rhamnopyranose, CALCIUM ION, ... | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
5YT3
 
 | | Structure of the Human Mitogen-Activated Protein Kinase Kinase 1 S218D and S222D mutant | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase kinase 1, isoform CRA_d, ... | | Authors: | Nakae, S, Doko, K, Tada, T, Shirai, T. | | Deposit date: | 2017-11-16 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Human Mitogen-Activated Protein Kinase Kinase 1 S218D and S222D mutant
To Be Published
|
|
5XQ3
 
 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum | | Descriptor: | CALCIUM ION, Pcrglx protein | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-06 | | Release date: | 2018-03-21 | | Last modified: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
6A9K
 
 | | Crystal structure of the complex of the hydrolytic antibody Fab 9C10 with a transition-state analog | | Descriptor: | 5-[(2R,3R)-2-[2,2-bis(chloranyl)ethanoylamino]-3-(4-nitrophenyl)-3-[oxidanyl-[[4-[2,2,2-tris(fluoranyl)ethanoylamino]phenyl]methyl]phosphoryl]oxy-propoxy]-5-oxidanylidene-pentanoic acid, IMMUNOGLOBULIN 9C10 H CHAIN, IMMUNOGLOBULIN 9C10 L CHAIN | | Authors: | Tsuchiya, Y, Fujii, I, Tada, T, Yamaguchi, A, Tsumuraya, T, Kumon, A. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of the hydrolytic antibody Fab 9C10 with a transition-state analog
To Be Published
|
|
6A8I
 
 | | Crystal structure of endo-arabinanase ABN-TS D147N mutant in complex with arabinohexaose | | Descriptor: | CALCIUM ION, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, endo-alpha-(1->5)-L-arabinanase | | Authors: | Yamaguchi, A, Tada, T. | | Deposit date: | 2018-07-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of endo-1,5-alpha-L-arabinanase mutants from Bacillus thermodenitrificans TS-3 in complex with arabino-oligosaccharides.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3W8Q
 
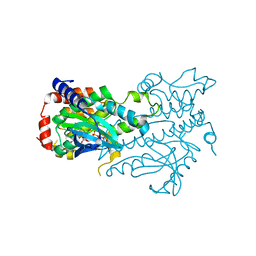 | | Structure of the Human Mitogen-Activated Protein Kinase Kinase 1 (MEK1) | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Nakae, S, Kitamura, M, Shirai, T, Tada, T. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of mitogen-activated protein kinase kinase 1 in the DFG-out conformation.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
1IYN
 
 | |
3W5F
 
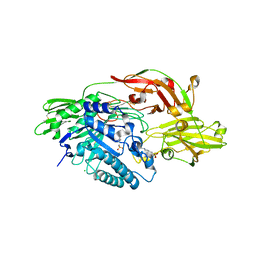 | | Crystal structure of tomato beta-galactosidase 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eda, M, Tada, T. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of tomato beta-galactosidase 4
To be Published
|
|
