6BA7
 
 | |
4PPR
 
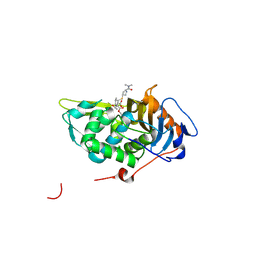 | | Crystal structure of Mycobacterium tuberculosis D,D-peptidase Rv3330 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein DacB1 | | Authors: | Prigozhin, D.M, Huizar, J.P, Mavrici, D, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-02-27 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subfamily-specific adaptations in the structures of two penicillin-binding proteins from Mycobacterium tuberculosis.
Plos One, 9, 2014
|
|
6AXB
 
 | |
6C9V
 
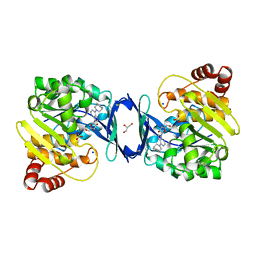 | | Mycobacterium tuberculosis adenosine kinase bound to (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-(4-phenylpiperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol | | Descriptor: | (2R,3S,4R,5R)-2-(hydroxymethyl)-5-[6-(4-phenylpiperazin-1-yl)-9H-purin-9-yl]tetrahydrofuran-3,4-diol, Adenosine kinase, GLYCEROL, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
6C9Q
 
 | |
6C9R
 
 | | Mycobacterium tuberculosis adenosine kinase bound to (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-(thiophen-3-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol | | Descriptor: | 9-beta-D-ribofuranosyl-6-(thiophen-3-yl)-9H-purine, Adenosine kinase, GLYCEROL, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
6C67
 
 | | Mycobacterium tuberculosis adenosine kinase bound to iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Adenosine kinase, GLYCEROL, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-17 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
6C6O
 
 | |
6C9S
 
 | | Mycobacterium tuberculosis adenosine kinase bound to (2R,3R,4S,5R)-2-(6-([1,1'-biphenyl]-4-ylethynyl)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol | | Descriptor: | 6-[([1,1'-biphenyl]-4-yl)ethynyl]-9-beta-D-ribofuranosyl-9H-purine, Adenosine kinase, SODIUM ION, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
6C9N
 
 | |
6C7B
 
 | |
6C8P
 
 | |
2ASF
 
 | | Crystal structure of the conserved hypothetical protein Rv2074 from Mycobacterium tuberculosis 1.6 A | | Descriptor: | CITRIC ACID, Hypothetical protein Rv2074, SODIUM ION | | Authors: | Biswal, B.K, Au, K, Cherney, M.M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The molecular structure of Rv2074, a probable pyridoxine 5'-phosphate oxidase from Mycobacterium tuberculosis, at 1.6 angstroms resolution.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
4P0M
 
 | | Crystal structure of an evolved putative penicillin-binding protein homolog, Rv2911, from Mycobacterium tuberculosis | | Descriptor: | D-alanyl-D-alanine carboxypeptidase | | Authors: | Krieger, I, Yu, M, Bursey, E, Hung, L.-W, Terwilliger, T.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subfamily-Specific Adaptations in the Structures of Two Penicillin-Binding Proteins from Mycobacterium tuberculosis.
Plos One, 9, 2014
|
|
6C2X
 
 | |
6C9P
 
 | |
3DBO
 
 | | Crystal structure of a member of the VapBC family of toxin-antitoxin systems, VapBC-5, from Mycobacterium tuberculosis | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, SODIUM ION, ... | | Authors: | Miallau, L, Cascio, D, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure and Proposed Activity of a Member of the VapBC Family of Toxin-Antitoxin Systems: VapBC-5 FROM MYCOBACTERIUM TUBERCULOSIS.
J.Biol.Chem., 284, 2009
|
|
3H6P
 
 | | Crystal structure of Rv3019c-Rv3020c from Mycobacterium tuberculosis | | Descriptor: | ESAT-6 LIKE PROTEIN ESXS, ESAT-6-like protein esxR, GLYCEROL | | Authors: | Chan, S, Arbing, M, Phan, T, Kaufmann, M, Cascio, D, Eisenberg, D, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-04-23 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of Rv3019c-Rv3020c from Mycobacterium tuberculosis
To be Published
|
|
4KBJ
 
 | |
3F69
 
 | | Crystal structure of the Mycobacterium tuberculosis PknB mutant kinase domain in complex with KT5720 | | Descriptor: | SULFATE ION, Serine/threonine-protein kinase pknB, hexyl (5S,6R,8R)-6-hydroxy-5-methyl-13-oxo-5,6,7,8-tetrahydro-13H-5,8-epoxy-4b,8a,14-triazadibenzo[b,h]cycloocta[1,2,3,4-jkl]c yclopenta[e]-as-indacene-6-carboxylate | | Authors: | Alber, T, Mieczkowski, C.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Auto-activation mechanism of the Mycobacterium tuberculosis PknB receptor Ser/Thr kinase.
Embo J., 27, 2008
|
|
1RFE
 
 | | Crystal structure of conserved hypothetical protein Rv2991 from Mycobacterium tuberculosis | | Descriptor: | hypothetical protein Rv2991 | | Authors: | Benini, S, Haouz, A, Proux, F, Betton, J.M, Alzari, P, Dodson, G.G, Wilson, K.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-11-08 | | Release date: | 2004-12-28 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Rv2991 from Mycobacterium tuberculosis: An F420binding protein with unknown function.
J. Struct. Biol., 2019
|
|
4M6G
 
 | | Structure of the Mycobacterium tuberculosis peptidoglycan amidase Rv3717 in complex with L-Alanine-iso-D-Glutamine reaction product | | Descriptor: | ALANINE, D-alpha-glutamine, Peptidoglycan Amidase Rv3717, ... | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
4M6H
 
 | | Structure of the reduced, metal-free form of Mycobacterium tuberculosis peptidoglycan amidase Rv3717 | | Descriptor: | Peptidoglycan Amidase Rv3717 | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
4KBM
 
 | |
4M6I
 
 | | Structure of the reduced, Zn-bound form of Mycobacterium tuberculosis peptidoglycan amidase Rv3717 | | Descriptor: | Peptidoglycan Amidase Rv3717, ZINC ION | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.666 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
