6UUY
 
 | |
2H0R
 
 | | Structure of the Yeast Nicotinamidase Pnc1p | | Descriptor: | Nicotinamidase, ZINC ION | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2006-05-15 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the yeast nicotinamidase Pnc1p.
Arch.Biochem.Biophys., 461, 2007
|
|
3RBZ
 
 | | MthK channel, Ca2+-bound | | Descriptor: | CALCIUM ION, Calcium-gated potassium channel mthK | | Authors: | Taylor, A.B, Parfenova, L.V, Rothberg, B.S. | | Deposit date: | 2011-03-30 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of multiple Ca2+-binding sites in a K+ channel RCK domain
Proc.Natl.Acad.Sci.USA, 2011
|
|
4OTA
 
 | |
4OTC
 
 | |
1ZPU
 
 | | Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (I) ION, ... | | Authors: | Taylor, A.B, Stoj, C.S, Ziegler, L, Kosman, D.J, Hart, P.J. | | Deposit date: | 2005-05-17 | | Release date: | 2005-10-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The copper-iron connection in biology: Structure of the metallo-oxidase Fet3p.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4GDP
 
 | | Yeast polyamine oxidase FMS1, N195A mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1, TETRAETHYLENE GLYCOL | | Authors: | Taylor, A.B, Adachi, M.S, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2012-08-01 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9998 Å) | | Cite: | Mechanistic and Structural Analyses of the Roles of Active Site Residues in Yeast Polyamine Oxidase Fms1: Characterization of the N195A and D94N Enzymes.
Biochemistry, 51, 2012
|
|
1HR6
 
 | | Yeast Mitochondrial Processing Peptidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HR8
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Cytochrome C Oxidase IV Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYTOCHROME C OXIDASE POLYPEPTIDE IV, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
4OTB
 
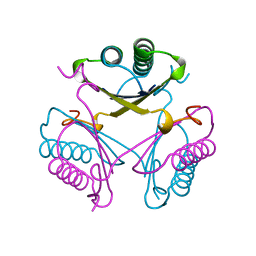 | |
1HR9
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Malate Dehydrogenase Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MALATE DEHYDROGENASE, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HR7
 
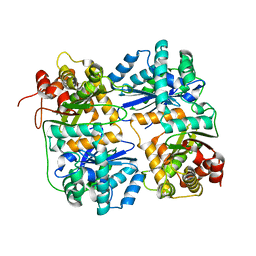 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant | | Descriptor: | MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ZINC ION | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
5U9M
 
 | |
5TX2
 
 | | Miniature TGF-beta2 3-mutant monomer | | Descriptor: | Transforming growth factor beta-2 | | Authors: | Taylor, A.B, Kim, S.K, Hart, P.J, Hinck, A.P. | | Deposit date: | 2016-11-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | An engineered transforming growth factor beta (TGF-beta ) monomer that functions as a dominant negative to block TGF-beta signaling.
J. Biol. Chem., 292, 2017
|
|
5UWK
 
 | | Matrix metalloproteinase-13 complexed with selective inhibitor compound (S)-10a | | Descriptor: | (S)-3-methyl-2-(4'-(((4-oxo-4,5,6,7-tetrahydro-3H-cyclopenta[d]pyrimidin-2-yl)thio)methyl)-[1,1'-biphenyl]-4-ylsulfonamido)butanoic acid, CALCIUM ION, Collagenase 3, ... | | Authors: | Taylor, A.B, Cao, X, Hart, P.J. | | Deposit date: | 2017-02-21 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective Matrix Metalloproteinase 13 Inhibitors.
J. Med. Chem., 60, 2017
|
|
5TIW
 
 | |
5T0R
 
 | |
5T0S
 
 | | Synaptotagmin 1 C2B domain, cadmium-bound | | Descriptor: | CADMIUM ION, SODIUM ION, Synaptotagmin-1 | | Authors: | Taylor, A.B, Hart, P.J, Igumenova, T.I. | | Deposit date: | 2016-08-16 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Non-Native Metal Ion Reveals the Role of Electrostatics in Synaptotagmin 1-Membrane Interactions.
Biochemistry, 56, 2017
|
|
4ECH
 
 | | Yeast Polyamine Oxidase FMS1, H67Q Mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1 | | Authors: | Taylor, A.B, Adachi, M.S, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2012-03-26 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic and structural analyses of the role of his67 in the yeast polyamine oxidase fms1.
Biochemistry, 51, 2012
|
|
5TIX
 
 | | Schistosoma haematobium (Blood Fluke) Sulfotransferase/R-oxamniquine Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase, {(2R)-7-nitro-2-[(propan-2-ylamino)methyl]-1,2,3,4-tetrahydroquinolin-6-yl}methanol | | Authors: | Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and enzymatic insights into species-specific resistance to schistosome parasite drug therapy.
J. Biol. Chem., 292, 2017
|
|
5TIV
 
 | | Schistosoma haematobium (Blood Fluke) Sulfotransferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase | | Authors: | Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural and enzymatic insights into species-specific resistance to schistosome parasite drug therapy.
J. Biol. Chem., 292, 2017
|
|
5TIY
 
 | | Schistosoma haematobium (Blood Fluke) Sulfotransferase/S-oxamniquine Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase, {(2S)-7-nitro-2-[(propan-2-ylamino)methyl]-1,2,3,4-tetrahydroquinolin-6-yl}methanol | | Authors: | Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and enzymatic insights into species-specific resistance to schistosome parasite drug therapy.
J. Biol. Chem., 292, 2017
|
|
5TIZ
 
 | | Schistosoma japonicum (Blood Fluke) Sulfotransferase | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase | | Authors: | Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural and enzymatic insights into species-specific resistance to schistosome parasite drug therapy.
J. Biol. Chem., 292, 2017
|
|
8E5R
 
 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase/CIDD-0150610 Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase oxamniquine resistance protein, [4-({[(3R)-1-[(1H-indol-3-yl)methyl]-3-{[4-(trifluoromethyl)phenyl]methyl}pyrrolidin-3-yl]methyl}amino)-3-nitrophenyl]methanol | | Authors: | Taylor, A.B, Alwan, S.N. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Oxamniquine derivatives overcome Praziquantel treatment limitations for Schistosomiasis.
Plos Pathog., 19, 2023
|
|
8E5Q
 
 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase/CIDD-0150303 Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase oxamniquine resistance protein, [2-(dihydroxyamino)-4-({[(3R)-1-[(1H-indol-3-yl)methyl]-3-{[3-(trifluoromethyl)phenyl]methyl}pyrrolidin-3-yl]methyl}amino)phenyl]methanol | | Authors: | Taylor, A.B, Alwan, S.N. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Oxamniquine derivatives overcome Praziquantel treatment limitations for Schistosomiasis.
Plos Pathog., 19, 2023
|
|
