5W3A
 
 | |
5W2K
 
 | |
5W2R
 
 | |
5W2X
 
 | |
5W31
 
 | |
5W3B
 
 | |
6BRT
 
 | | F-box protein CTH with hydrolase | | Descriptor: | (5R)-5-hydroxy-3-methylfuran-2(5H)-one, D3-CTH-D14-D-ring | | Authors: | Shabek, N, Zheng, N, Mao, H, Hinds, T.R, Ticchiarelli, F, Leyser, O. | | Deposit date: | 2017-12-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Nature, 563, 2018
|
|
5W70
 
 | | X-ray Structure of RbmB from Streptomyces ribosidificus | | Descriptor: | 1,2-ETHANEDIOL, L-glutamine:2-deoxy-scyllo-inosose aminotransferase, [4-({[(1R,2S,3S,4R,5S)-5-amino-2,3,4-trihydroxycyclohexyl]amino}methyl)-5-hydroxy-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Zachman-Brockmeyer, T.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of RbmB from Streptomyces ribosidificus, an aminotransferase involved in the biosynthesis of ribostamycin.
Protein Sci., 26, 2017
|
|
5W2D
 
 | |
5W30
 
 | | Crystal structure of mutant CJ YCEI protein (CJ-N48C) with monobromobimane guest structure | | Descriptor: | 3-(bromomethyl)-2,5,6-trimethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, Putative periplasmic protein, SULFATE ION, ... | | Authors: | Huber, T.R, Snow, C.D. | | Deposit date: | 2017-06-07 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Installing Guest Molecules at Specific Sites within Scaffold Protein Crystals.
Bioconjug. Chem., 29, 2018
|
|
5VL5
 
 | | Coordination Chemistry within a Protein Host: Regulation of the Secondary Coordination Sphere | | Descriptor: | COPPER (II) ION, Streptavidin, [N-(3-{bis[2-(pyridin-2-yl-kappaN)ethyl]amino-kappaN}propyl)-5-(2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamide](azido)(hydroxy)copper | | Authors: | Mann, S.I, Heinisch, T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2017-04-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Coordination chemistry within a protein host: regulation of the secondary coordination sphere.
Chem. Commun. (Camb.), 54, 2018
|
|
5UF4
 
 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) O13 with LNnT bound | | Descriptor: | O13, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Gunn, R.J, Collins, B.C, McKitrick, T.R, Cummings, R.D, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-01-03 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
5W37
 
 | |
8P2W
 
 | | Structure of human SIT1 (focussed map / refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Sodium- and chloride-dependent transporter XTRP3 | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P2Z
 
 | | Structure of human SIT1 bound to L-pipecolate (focussed map / refinement) | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P30
 
 | | Structure of human SIT1:ACE2 complex (open PD conformation) bound to L-pipecolate | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
6US6
 
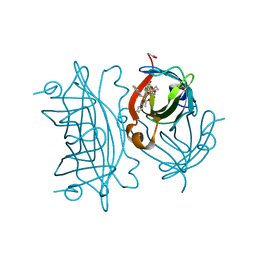 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | ACETATE ION, Streptavidin, {N-(2-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}iron(3+) | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
5WBB
 
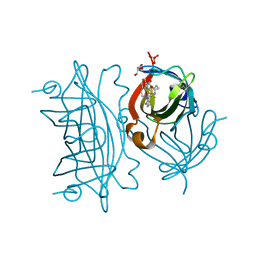 | | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins - S112A | | Descriptor: | COPPER (II) ION, GLYCEROL, SULFATE ION, ... | | Authors: | Mann, S.I, Heinisch, T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
6UIZ
 
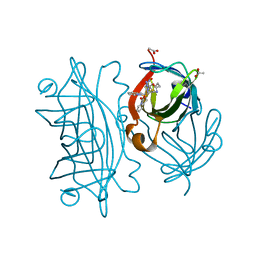 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | ACETATE ION, Streptavidin, {N-(2-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(triaza-1,2-dien-2-ium-1-ide-kappaN~1~)iron(4+) | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
6UI0
 
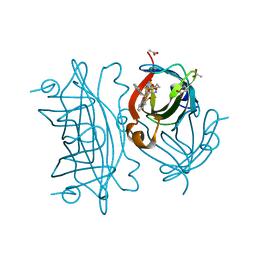 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | ACETATE ION, Streptavidin, {N-(2-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}iron(3+) | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-09-29 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
5WBA
 
 | | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins - WT | | Descriptor: | ACETATE ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | Mann, S.I, Heinisch, T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
8OJW
 
 | |
8OJX
 
 | | Streptavidin S112YK121E artificial metalloenzyme for carboamination | | Descriptor: | IRIDIUM ION, Streptavidin, trichloro{(1,2,3,4,5-eta)-1,2,3,4-tetramethyl-5-[2-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)ethyl]cyclopentadienyl}rhodium(1+) | | Authors: | Igareta, N.V, Ward, T.R. | | Deposit date: | 2023-03-25 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Streptavidin WT artificial metalloenzyme for carboamination
To Be Published
|
|
6BRP
 
 | | F-box protein form 2 | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Shabek, N, Zheng, N, Mao, H, Hinds, T.R, Ticchiarelli, F, Leyser, O. | | Deposit date: | 2017-11-30 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Nature, 563, 2018
|
|
5WBC
 
 | | Designed Artificial Cupredoxins - WT | | Descriptor: | ACETATE ION, GLYCEROL, Streptavidin, ... | | Authors: | Mann, S.I, Heinisch, T, Weitz, A.C, Hendrich, M.R, Ward, T.R, Borovik, A.S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Modular Artificial Cupredoxins.
J. Am. Chem. Soc., 138, 2016
|
|
