1FL1
 
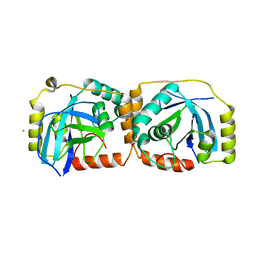 | | KSHV PROTEASE | | Descriptor: | POTASSIUM ION, PROTEASE | | Authors: | Reiling, K.K, Pray, T.R, Craik, C.S, Stroud, R.M. | | Deposit date: | 2000-08-11 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional consequences of the Kaposi's sarcoma-associated herpesvirus protease structure: regulation of activity and dimerization by conserved structural elements.
Biochemistry, 39, 2000
|
|
1FA9
 
 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE A COMPLEXED WITH AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCOGEN PHOSPHORYLASE, LIVER FORM, ... | | Authors: | Rath, V.L, Ammirati, M, LeMotte, P.K, Fennell, K.F, Mansour, M.N, Danley, D.E, Hynes, T.R, Schulte, G.K, Wasilko, D.J, Pandit, J. | | Deposit date: | 2000-07-12 | | Release date: | 2000-08-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation of human liver glycogen phosphorylase by alteration of the secondary structure and packing of the catalytic core.
Mol.Cell, 6, 2000
|
|
1FGM
 
 | |
1GID
 
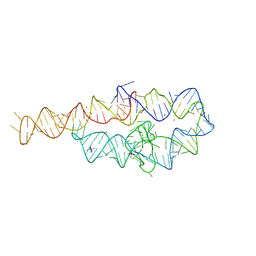 | | CRYSTAL STRUCTURE OF A GROUP I RIBOZYME DOMAIN: PRINCIPLES OF RNA PACKING | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, P4-P6 RNA RIBOZYME DOMAIN | | Authors: | Cate, J.H, Gooding, A.R, Podell, E, Zhou, K, Golden, B.L, Kundrot, C.E, Cech, T.R, Doudna, J.A. | | Deposit date: | 1996-08-22 | | Release date: | 1996-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a group I ribozyme domain: principles of RNA packing.
Science, 273, 1996
|
|
1GL2
 
 | | Crystal structure of an endosomal SNARE core complex | | Descriptor: | ENDOBREVIN, SYNTAXIN 7, SYNTAXIN 8, ... | | Authors: | Antonin, W, Becker, S, Jahn, R, Schneider, T.R. | | Deposit date: | 2001-08-22 | | Release date: | 2002-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Endosomal Snare Complex Reveals Common Structural Principles of All Snares.
Nat.Struct.Biol., 9, 2001
|
|
5W3A
 
 | |
1GRZ
 
 | |
5W71
 
 | | X-ray structure of BtrR from Bacillus circulans in the presence of the 2-DOS external aldimine | | Descriptor: | CHLORIDE ION, L-glutamine:2-deoxy-scyllo-inosose aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Zachman-Brockmeyer, T.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of RbmB from Streptomyces ribosidificus, an aminotransferase involved in the biosynthesis of ribostamycin.
Protein Sci., 26, 2017
|
|
5W2Z
 
 | |
5W32
 
 | |
5W3C
 
 | |
5W2K
 
 | |
5W2R
 
 | |
5W2X
 
 | |
5W31
 
 | |
5W3B
 
 | |
5W70
 
 | | X-ray Structure of RbmB from Streptomyces ribosidificus | | Descriptor: | 1,2-ETHANEDIOL, L-glutamine:2-deoxy-scyllo-inosose aminotransferase, [4-({[(1R,2S,3S,4R,5S)-5-amino-2,3,4-trihydroxycyclohexyl]amino}methyl)-5-hydroxy-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Zachman-Brockmeyer, T.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of RbmB from Streptomyces ribosidificus, an aminotransferase involved in the biosynthesis of ribostamycin.
Protein Sci., 26, 2017
|
|
1HB0
 
 | | Snapshots of serine protease catalysis: (D) acyl-enzyme intermediate between porcine pancreatic elastase and human beta-casomorphin-7 jumped to pH 10 for 2 minutes | | Descriptor: | CALCIUM ION, ELASTASE 1, SULFATE ION | | Authors: | Wilmouth, R.C, Edman, K, Neutze, R, Wright, P.A, Clifton, I.J, Schneider, T.R, Schofield, C.J, Hajdu, J. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-Ray Snapshots of Serine Protease Catalysis Reveal a Tetrahedral Intermediate
Nat.Struct.Biol., 8, 2001
|
|
1HAY
 
 | | Snapshots of serine protease catalysis: (B) acyl-enzyme intermediate between porcine pancreatic elastase and human beta-casomorphin-7 jumped to pH 10 for 10 seconds | | Descriptor: | CALCIUM ION, ELASTASE 1, SULFATE ION | | Authors: | Wilmouth, R.C, Edman, K, Neutze, R, Wright, P.A, Clifton, I.J, Schneider, T.R, Schofield, C.J, Hajdu, J. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-Ray Snapshots of Serine Protease Catalysis Reveal a Tetrahedral Intermediate
Nat.Struct.Biol., 8, 2001
|
|
5ZAT
 
 | | Crystal structure of 5-carboxylcytosine containing decamer dsDNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*(CAC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
1HFB
 
 | |
1HAX
 
 | | Snapshots of serine protease catalysis: (A) acyl-enzyme intermediate between porcine pancreatic elastase and human beta-casomorphin-7 at pH 5 | | Descriptor: | BETA-CASOMORPHIN-7, CALCIUM ION, ELASTASE 1, ... | | Authors: | Wilmouth, R.C, Edman, K, Neutze, R, Wright, P.A, Clifton, I.J, Schneider, T.R, Schofield, C.J, Hajdu, J. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Snapshots of Serine Protease Catalysis Reveal a Tetrahedral Intermediate
Nat.Struct.Biol., 8, 2001
|
|
5ZAS
 
 | | Crystal structure of 5-formylcytosine containing decamer dsDNA | | Descriptor: | BICARBONATE ION, DNA (5'-D(*CP*CP*AP*GP*(5FC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
1G5N
 
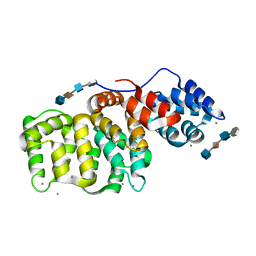 | | ANNEXIN V COMPLEX WITH HEPARIN OLIGOSACCHARIDES | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ANNEXIN V, CALCIUM ION | | Authors: | Capila, I, Heraiz, M.J, Mo, Y.D, Mealy, T.R, Campos, B, Dedman, J.R, Linhardt, R.J, Seaton, B.A. | | Deposit date: | 2000-11-01 | | Release date: | 2001-06-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Annexin V--heparin oligosaccharide complex suggests heparan sulfate--mediated assembly on cell surfaces.
Structure, 9, 2001
|
|
1EKA
 
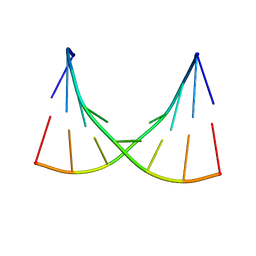 | | NMR AND MOLECULAR MODELING REVEAL THAT DIFFERENT HYDROGEN BONDING PATTERNS ARE POSSIBLE FOR GU PAIRS: ONE HYDROGEN BOND FOR EACH GU PAIR IN R(GGCGUGCC)2 AND TWO FOR EACH GU PAIR IN R(GAGUGCUC)2 | | Descriptor: | RNA (5'-R(*GP*AP*GP*UP*GP*CP*UP*C)-3') | | Authors: | Chen, X, McDowell, J.A, Kierzek, R, Krugh, T.R, Turner, D.H. | | Deposit date: | 2000-03-07 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2).
Biochemistry, 39, 2000
|
|
