1S1D
 
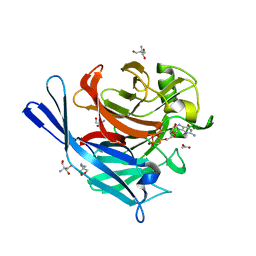 | | Structure and protein design of human apyrase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Dai, J, Liu, J, Deng, Y, Smith, T.M, Lu, M. | | Deposit date: | 2004-01-06 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and protein design of a human platelet function inhibitor.
Cell(Cambridge,Mass.), 116, 2004
|
|
1R5K
 
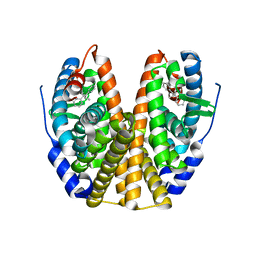 | | Human Estrogen Receptor alpha Ligand-Binding Domain In Complex With GW5638 | | Descriptor: | (2E)-3-{4-[(1E)-1,2-DIPHENYLBUT-1-ENYL]PHENYL}ACRYLIC ACID, Estrogen receptor | | Authors: | Wu, Y.-L, Yang, X, Ren, Z, McDonnell, D.P, Norris, J.D, Willson, T.M, Greene, G.L. | | Deposit date: | 2003-10-10 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for an unexpected mode of SERM-mediated ER antagonism.
Mol.Cell, 18, 2005
|
|
3NRX
 
 | | Insights into anti-parallel microtubule crosslinking by PRC1, a conserved non-motor microtubule binding protein | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Kapoor, T.M, Subramanian, R, Wilson-Kubalek, E.M, Arthur, C.P, Bick, M.J, Campbell, E.A, Darst, S.A, Milligan, R.A. | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into Antiparallel Microtubule Crosslinking by PRC1, a Conserved Nonmotor Microtubule Binding Protein.
Cell(Cambridge,Mass.), 142, 2010
|
|
3NYZ
 
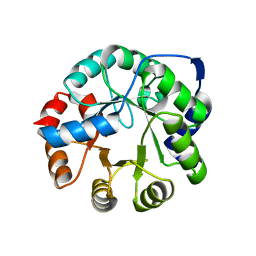 | | Crystal Structure of Kemp Elimination Catalyst 1A53-2 | | Descriptor: | Indole-3-glycerol phosphate synthase, SULFATE ION | | Authors: | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.514 Å) | | Cite: | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1U6O
 
 | | Mispairing of a Site-Specific Major Groove (2S,3S)-N6-(2,3,4-Trihydroxybutyl)-2-deoxyadenosyl DNA Adduct of Butadiene Diol Epoxide with Deoxyguanosine: Formation of a dA(anti)dG(anti) Pairing Interaction | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(2BU)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*GP*GP*TP*CP*CP*G)-3' | | Authors: | Scholdberg, T.A, Nechev, L.N, Merritt, W.K, Harris, T.M, Harris, C.M, Lloyd, R.S, Stone, M.P. | | Deposit date: | 2004-07-30 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mispairing of a Site Specific Major Groove (2S,3S)-N6-(2,3,4-Trihydroxybutyl)-2'-deoxyadenosyl DNA Adduct of Butadiene Diol Epoxide with Deoxyguanosine: Formation of a dA(anti)dG(anti) Pairing Interaction.
CHEM.RES.TOXICOL., 18, 2005
|
|
1U6C
 
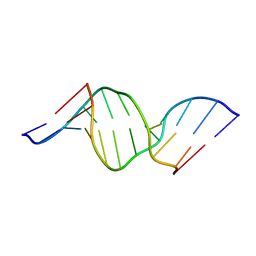 | | The NMR-derived solution structure of the (2S,3S)-N6-(2,3,4-trihydroxybutyl)-2'-deoxyadenosyl DNA adduct of butadiene diol epoxide | | Descriptor: | (2S,3S)-N6-(2,3,4-trihydroxybutyl)-2'-deoxyadenosyl DNA adduct of butadiene diol epoxide | | Authors: | Scholdberg, T.A, Nechev, L.N, Merritt, W.K, Harris, T.M, Harris, C.M, Lloyd, R.S, Stone, M.P. | | Deposit date: | 2004-07-29 | | Release date: | 2004-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a site specific major groove (2S,3S)-N6-(2,3,4-trihydroxybutyl)-2'-deoxyadenosyl DNA adduct of butadiene diol epoxide.
Chem.Res.Toxicol., 17, 2004
|
|
3M8Y
 
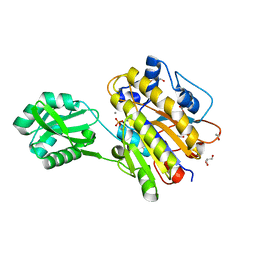 | | Phosphopentomutase from Bacillus cereus after glucose-1,6-bisphosphate activation | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Panosian, T.D, Nannemann, D.P, Watkins, G, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2010-03-19 | | Release date: | 2010-12-29 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
1S3K
 
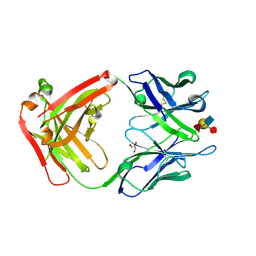 | | Crystal Structure of a Humanized Fab (hu3S193) in Complex with the Lewis Y Tetrasaccharide | | Descriptor: | GLYCEROL, HU3S193 Fab fragment, heavy chain, ... | | Authors: | Ramsland, P.A, Farrugia, W, Bradford, T.M, Hogarth, P.M, Scott, A.M. | | Deposit date: | 2004-01-13 | | Release date: | 2004-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural convergence of antibody binding of carbohydrate determinants in lewis y tumor antigens
J.Mol.Biol., 340, 2004
|
|
1T2T
 
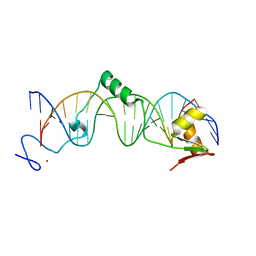 | | Crystal structure of the DNA-binding domain of intron endonuclease I-TevI with operator site | | Descriptor: | 5'-D(*AP*AP*TP*TP*AP*AP*AP*GP*GP*GP*CP*AP*GP*TP*CP*CP*TP*AP*CP*AP*A)-3', 5'-D(*TP*TP*TP*GP*TP*AP*GP*GP*AP*CP*TP*GP*CP*CP*CP*TP*TP*TP*AP*AP*T)-3', Intron-associated endonuclease 1, ... | | Authors: | Edgell, D.R, Derbyshire, V, Van Roey, P, LaBonne, S, Stanger, M.J, Li, Z, Boyd, T.M, Shub, D.A, Belfort, M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intron-encoded homing endonuclease I-TevI also functions as a transcriptional autorepressor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3M8W
 
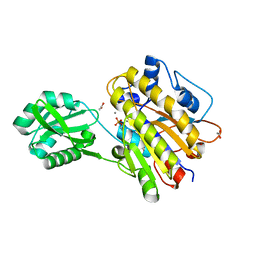 | | Phosphopentomutase from Bacillus cereus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Panosian, T.D, Nannemann, D.P, Watkins, G, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2010-03-19 | | Release date: | 2010-12-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3NYD
 
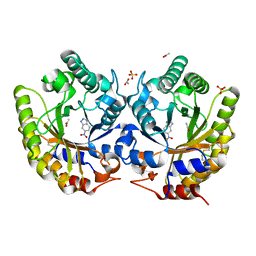 | | Crystal Structure of Kemp Eliminase HG-2 Complexed with Transition State Analog 5-Nitro Benzotriazole | | Descriptor: | 5-nitro-1H-benzotriazole, ACETATE ION, Endo-1,4-beta-xylanase, ... | | Authors: | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | Deposit date: | 2010-07-14 | | Release date: | 2011-06-29 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3OAA
 
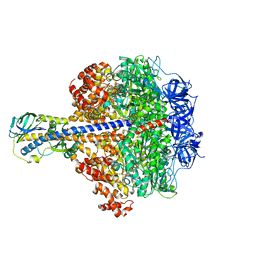 | |
1U33
 
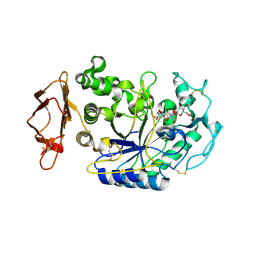 | | In situ extension as an approach for identifying novel alpha-amylase inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4'-O-METHYL-MALTOSYL-ALPHA (1,4)-(Z, 3S,4S,5R,6R)-3,4,5-TRIHYDROXY-6-HYDROXYMETHYL-PIPERIDIN-2-ONE, ... | | Authors: | Numao, S, Li, C, Damager, I, Wrodnigg, T.M, Begum, A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Situ Extension as an Approach for Identifying Novel alpha-Amylase Inhibitors.
J.Biol.Chem., 279, 2004
|
|
1U30
 
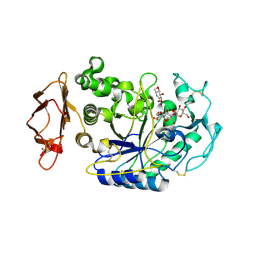 | | In situ extension as an approach for identifying novel alpha-amylase inhibitors, structure containing maltosyl-alpha (1,4)-D-gluconhydroximo-1,5-lactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Numao, S, Li, C, Damager, I, Wrodnigg, T.M, Begum, A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In Situ Extension as an Approach for Identifying Novel alpha-Amylase Inhibitors.
J.Biol.Chem., 279, 2004
|
|
1U6N
 
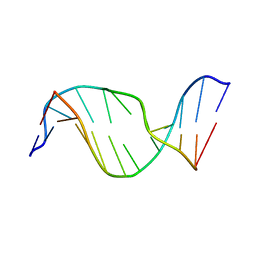 | | Solution Structure of an Oligodeoxynucleotide Containing a Butadiene Derived N1 b-Hydroxyalkyl Adduct on Deoxyinosine in the Human N-ras Codon 61 Sequence | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(2BD)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Scholdberg, T.A, Merritt, W.K, Dean, S.M, Kowalcyzk, A, Harris, T.M, Harris, C.M, Lloyd, R.S, Stone, M.P. | | Deposit date: | 2004-07-30 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an Oligodeoxynucleotide Containing a Butadiene Oxide-Derived N1 Beta-Hydroxyalkyl Deoxyinosine Adduct in the Human N-ras Codon 61 Sequence.
Biochemistry, 44, 2005
|
|
3M8Z
 
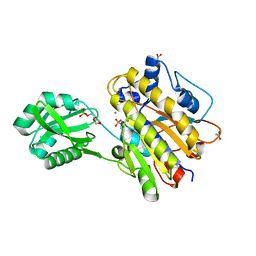 | | Phosphopentomutase from Bacillus cereus bound with ribose-5-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-alpha-D-ribofuranose, ACETATE ION, ... | | Authors: | Panosian, T.D, Nannemann, D.P, Watkins, G, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2010-03-19 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
1SEV
 
 | | Mature and translocatable forms of glyoxysomal malate dehydrogenase have different activities and stabilities but similar crystal structures | | Descriptor: | Malate dehydrogenase, glyoxysomal precursor | | Authors: | Cox, B.R, Chit, M.M, Weaver, T.M, Bailey, J, Gietl, C, Bell, E, Banaszak, L.J. | | Deposit date: | 2004-02-18 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Organelle and translocatable forms of glyoxysomal malate dehydrogenase. The effect of the N-terminal presequence
Febs J., 272, 2005
|
|
1S5L
 
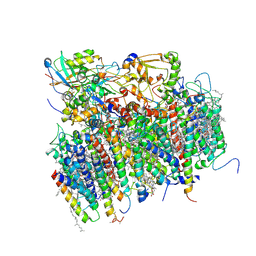 | | Architecture of the photosynthetic oxygen evolving center | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, BICARBONATE ION, ... | | Authors: | Ferreira, K.N, Iverson, T.M, Maghlaoui, K, Barber, J, Iwata, S. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-24 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Architecture of the Photosynthetic Oxygen-Evolving Center
Science, 303, 2004
|
|
7T2X
 
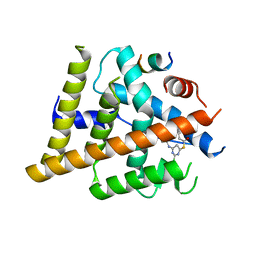 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S in Complex with 2-chloro-4-((4-hydroxybenzyl)amino)-5-phenylthieno[2,3-d]pyrimidin-6-ol and GRIP Peptide | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, S-(2-chloro-6-{[(4-hydroxyphenyl)methyl]amino}pyrimidin-4-yl) phenylethanethioate | | Authors: | Joiner, C, Sammeta, V.K.R, Norris, J.D, McDonnell, D.P, Wilson, T.M, Fanning, S.W. | | Deposit date: | 2021-12-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A New Chemotype of Chemically Tractable Nonsteroidal Estrogens Based on a Thieno[2,3- d ]pyrimidine Core.
Acs Med.Chem.Lett., 13, 2022
|
|
1RMN
 
 | | A THREE-DIMENSIONAL MODEL FOR THE HAMMERHEAD RIBOZYME BASED ON FLUORESCENCE MEASUREMENTS | | Descriptor: | RNA (49-MER) | | Authors: | Tuschl, T, Gohlke, C, Jovin, T.M, Westhof, E, Eckstein, F. | | Deposit date: | 1994-10-20 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | FLUORESCENCE TRANSFER | | Cite: | A three-dimensional model for the hammerhead ribozyme based on fluorescence measurements.
Science, 266, 1994
|
|
3N4I
 
 | | Crystal structure of the SHV-1 D104E beta-lactamase/beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Hanes, M.S, Reynolds, K.A, McNamara, C, Ghosh, P, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2010-05-21 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Specificity and Cooperativity at beta-lactamase Position 104 in TEM-1/BLIP and SHV-1/BLIP interactions
Proteins, 79, 2011
|
|
1T3P
 
 | | Half-sandwich arene ruthenium(II)-enzyme complex | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | McNae, I.W, Fishburne, K, Habtemariam, A, Hunter, T.M, Melchart, M, Wang, F, Walkinshaw, M.D, Sadler, P.J. | | Deposit date: | 2004-04-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Half-sandwich arene ruthenium(II)-enzyme complex
CHEM.COMMUN.(CAMB.), 16, 2004
|
|
1UWR
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with 2-deoxy-2-fluoro-galactose | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-galactopyranose, ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Studies of the Beta-Glycosidase from Sulfolobus Solfataricus in Complex with Covalently and Noncovalently Bound Inhibitors.
Biochemistry, 43, 2004
|
|
3NRY
 
 | | Insights into anti-parallel microtubule crosslinking by PRC1, a conserved microtubule binding protein | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Kapoor, T.M, Subramanian, R, Wilson-Kubalek, E.M, Arthur, C.P, Bick, M.J, Campbell, E.A, Darst, S.A, Milligan, R.A. | | Deposit date: | 2010-07-01 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into Antiparallel Microtubule Crosslinking by PRC1, a Conserved Nonmotor Microtubule Binding Protein.
Cell(Cambridge,Mass.), 142, 2010
|
|
1RSF
 
 | |
