2BCU
 
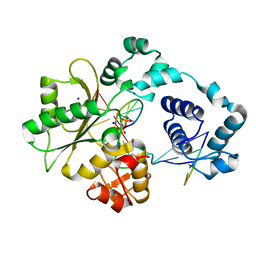 | | DNA polymerase lambda in complex with a DNA duplex containing an unpaired Damp and a T:T mismatch | | 分子名称: | 5'-D(*CP*AP*GP*TP*TP*CP*G)-3', 5'-D(*CP*GP*GP*CP*CP*GP*AP*TP*AP*CP*TP*G)-3', 5'-D(P*GP*CP*CP*G)-3', ... | | 著者 | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | 登録日 | 2005-10-19 | | 公開日 | 2006-03-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase
Cell(Cambridge,Mass.), 124, 2006
|
|
2BCS
 
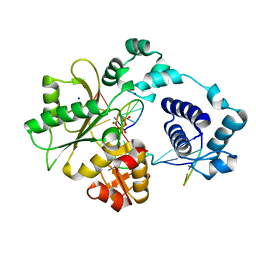 | | DNA polymerase lambda in complex with a DNA duplex containing an unpaired Dcmp | | 分子名称: | 5'-D(*CP*AP*GP*TP*AP*CP*G)-3', 5'-D(*CP*GP*GP*CP*CP*GP*CP*TP*AP*CP*TP*G)-3', 5'-D(*GP*CP*CP*G)-3', ... | | 著者 | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | 登録日 | 2005-10-19 | | 公開日 | 2006-03-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase
Cell(Cambridge,Mass.), 124, 2006
|
|
4M4W
 
 | |
2AS9
 
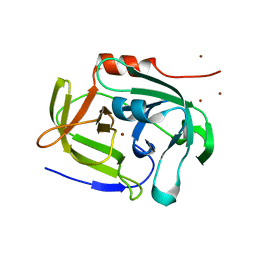 | | Functional and structural characterization of Spl proteases from staphylococcus aureus | | 分子名称: | ZINC ION, serine protease | | 著者 | Popowicz, G.M, Dubin, G, Stec-Niemczyk, J, Czarny, A, Dubin, A, Potempa, J, Holak, T.A. | | 登録日 | 2005-08-23 | | 公開日 | 2005-09-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Functional and Structural Characterization of Spl Proteases from Staphylococcus aureus
J.Mol.Biol., 358, 2006
|
|
1B77
 
 | |
1BDN
 
 | |
4LZD
 
 | | Human DNA polymerase mu- Apoenzyme | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-directed DNA/RNA polymerase mu, ... | | 著者 | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | 登録日 | 2013-07-31 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.849 Å) | | 主引用文献 | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1BRD
 
 | | Model for the structure of Bacteriorhodopsin based on high-resolution Electron Cryo-microscopy | | 分子名称: | BACTERIORHODOPSIN PRECURSOR, RETINAL | | 著者 | Henderson, R, Baldwin, J.M, Ceska, T.A, Zemlin, F, Beckmann, E, Downing, K.H. | | 登録日 | 1990-05-23 | | 公開日 | 1991-04-15 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | 主引用文献 | Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy.
J.Mol.Biol., 213, 1990
|
|
1CEJ
 
 | | SOLUTION STRUCTURE OF AN EGF MODULE PAIR FROM THE PLASMODIUM FALCIPARUM MEROZOITE SURFACE PROTEIN 1 | | 分子名称: | PROTEIN (MEROZOITE SURFACE PROTEIN 1) | | 著者 | Morgan, W.D, Birdsall, B, Frenkiel, T.A, Gradwell, M.G, Burghaus, P.A, Syed, S.E.H, Uthaipibull, C, Holder, A.A, Feeney, J. | | 登録日 | 1999-03-08 | | 公開日 | 1999-05-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an EGF module pair from the Plasmodium falciparum merozoite surface protein 1.
J.Mol.Biol., 289, 1999
|
|
2FBL
 
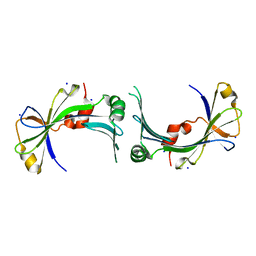 | | The crystal structure of the hypothetical protein NE1496 | | 分子名称: | SODIUM ION, hypothetical protein NE1496 | | 著者 | Lunin, V.V, Skarina, T, Onopriyenko, O, Binkowski, T.A, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-12-09 | | 公開日 | 2005-12-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The crystal structure of the hypothetical protein NE1496
To be Published
|
|
4LZG
 
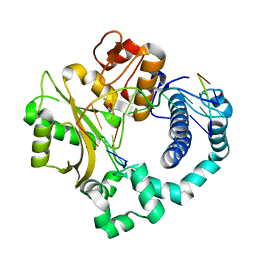 | | Binary complex of human DNA Polymerase Mu with DNA | | 分子名称: | CHLORIDE ION, DNA-directed DNA/RNA polymerase mu, GLYCEROL, ... | | 著者 | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | 登録日 | 2013-07-31 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1CLQ
 
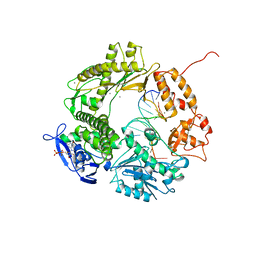 | |
1QSL
 
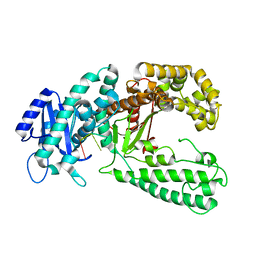 | |
1MRP
 
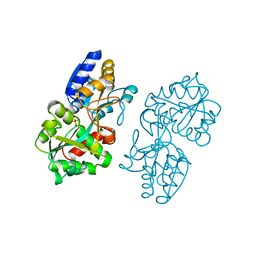 | | FERRIC-BINDING PROTEIN FROM HAEMOPHILUS INFLUENZAE | | 分子名称: | FE (III) ION, FERRIC IRON BINDING PROTEIN, PHOSPHATE ION | | 著者 | Bruns, C.M, Nowalk, A.J, Arvai, A.S, Mctigue, M.A, Vaughan, K.G, Mietzner, T.A, Mcree, D.E. | | 登録日 | 1997-05-14 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of Haemophilus influenzae Fe(+3)-binding protein reveals convergent evolution within a superfamily.
Nat.Struct.Biol., 4, 1997
|
|
4M0A
 
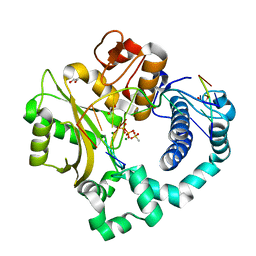 | | Human DNA Polymerase Mu post-catalytic complex | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA-directed DNA/RNA polymerase mu, ... | | 著者 | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | 登録日 | 2013-08-01 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4MDQ
 
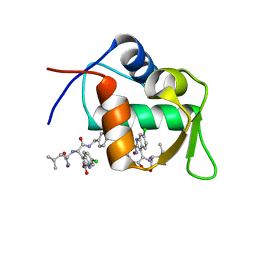 | | Structure of a novel submicromolar MDM2 inhibitor | | 分子名称: | 3-[(1R)-2-(benzylamino)-1-{[(2S)-1-(hydroxyamino)-4-methyl-1-oxopentan-2-yl]amino}-2-oxoethyl]-6-chloro-N-hydroxy-1H-indole-2-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Bista, M, Popowicz, G, Holak, T.A. | | 登録日 | 2013-08-23 | | 公開日 | 2013-11-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.119 Å) | | 主引用文献 | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
2DSM
 
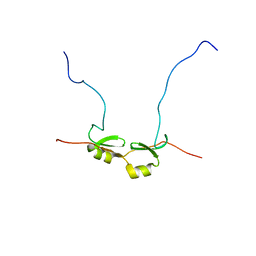 | | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450 | | 分子名称: | Hypothetical protein yqaI | | 著者 | Ramelot, T.A, Cort, J.R, Wang, D, Janua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-07-01 | | 公開日 | 2006-08-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450
to be published
|
|
2F1E
 
 | | Solution structure of ApaG protein | | 分子名称: | Protein apaG | | 著者 | Contessa, G, Pertinhez, T.A, Spisni, A, Paci, M, Farah, C.S, Cicero, D.O. | | 登録日 | 2005-11-14 | | 公開日 | 2006-10-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of ApaG from Xanthomonas axonopodis pv. citri reveals a fibronectin-3 fold.
Proteins, 67, 2007
|
|
4V9R
 
 | | Crystal structure of antibiotic DITYROMYCIN bound to 70S ribosome | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | 著者 | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | 登録日 | 2013-12-05 | | 公開日 | 2014-07-09 | | 最終更新日 | 2014-12-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
1GUB
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | 分子名称: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, NICKEL (II) ION | | 著者 | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | 登録日 | 2002-01-24 | | 公開日 | 2003-03-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
1QVG
 
 | | Structure of CCA oligonucleotide bound to the tRNA binding sites of the large ribosomal subunit of Haloarcula marismortui | | 分子名称: | 23S ribosomal rna, 50S RIBOSOMAL PROTEIN L10E, 50S ribosomal protein L13P, ... | | 著者 | Schmeing, T.M, Moore, P.B, Steitz, T.A. | | 登録日 | 2003-08-27 | | 公開日 | 2003-11-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit
RNA, 9, 2003
|
|
1GUD
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | 分子名称: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, ZINC ION | | 著者 | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | 登録日 | 2002-01-24 | | 公開日 | 2003-03-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
1Q86
 
 | | Crystal structure of CCA-Phe-cap-biotin bound simultaneously at half occupancy to both the A-site and P-site of the the 50S ribosomal Subunit. | | 分子名称: | 23S ribosomal rna, 50S ribosomal protein L13P, 50S ribosomal protein L14P, ... | | 著者 | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | 登録日 | 2003-08-20 | | 公開日 | 2003-10-07 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural insights into peptide bond formation.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1QJW
 
 | | CEL6A (Y169F) WITH A NON-HYDROLYSABLE CELLOTETRAOSE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, CELLOBIOHYDROLASE CEL6A (FORMERLY CALLED CBH II), ... | | 著者 | Zou, J.-Y, Jones, T.A. | | 登録日 | 1999-07-06 | | 公開日 | 1999-09-18 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystallographic Evidence for Substrate Ring Distortion and Protein Conformational Changes During Catalysis in Cellobiohydrolase Cel6A from Trichoderma Reesei
Structure, 7, 1999
|
|
1Q7Y
 
 | | Crystal Structure of CCdAP-Puromycin bound at the Peptidyl transferase center of the 50S ribosomal subunit | | 分子名称: | 23S ribosomal rna, 50S ribosomal protein L13P, 50S ribosomal protein L14P, ... | | 著者 | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | 登録日 | 2003-08-20 | | 公開日 | 2003-10-07 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural Insights Into Peptide Bond Formation
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
