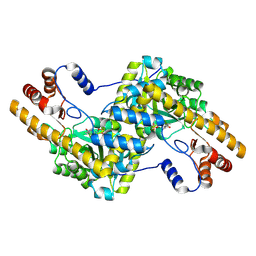
4F5H
 
 | |
2GM2
 
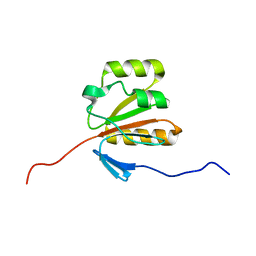 | | NMR structure of Xanthomonas campestris XCC1710: Northeast Structural Genomics Consortium target XcR35 | | Descriptor: | conserved hypothetical protein | | Authors: | Cort, J.R, Xiao, R, Wang, D.Y, Ma, L.C, Ciano, M, Montelione, G.T, Ramelot, T.A, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Xanthomonas campestris XCC1710 protein
To be Published
|
|
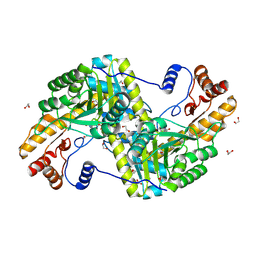
4F5L
 
 | |
5U42
 
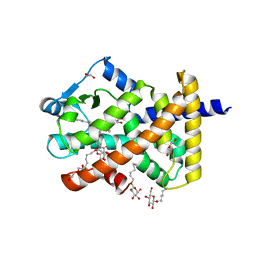 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 11 | | Descriptor: | 6-(2-{[cyclopropyl(3'-methoxy[1,1'-biphenyl]-4-carbonyl)amino]methyl}phenoxy)hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U40
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 15 | | Descriptor: | 6-[2-({benzyl[4-(furan-3-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TWR
 
 | | Pre-catalytic ternary complex of human Polymerase Mu (H329A) mutant with incoming nonhydrolyzable UMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2016-11-14 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
4G1M
 
 | | Re-refinement of alpha V beta 3 structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Springer, T.A, Mi, L, Zhu, J. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Alpha V Beta 3 Integrin Crystal Structures and their Functional Implications
Biochemistry, 51, 2012
|
|
5U3Z
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 10 | | Descriptor: | 6-[2-({cyclopropyl[4-(furan-3-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2HJQ
 
 | | NMR Structure of Bacillus Subtilis Protein YqbF, Northeast Structural Genomics Target SR449 | | Descriptor: | Hypothetical protein yqbF | | Authors: | Ding, K, Ramelot, T.A, Cort, J.R, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-30 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus Subtilis Protein YqbF, Northeast Structural Genomics Target SR449
TO BE PUBLISHED
|
|
5U3V
 
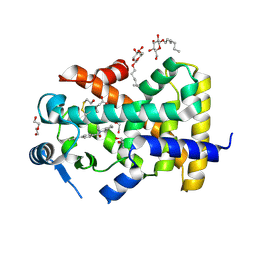 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 6 | | Descriptor: | 6-[2-({cyclopentyl[4-(furan-3-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7PEO
 
 | | Structure of the Caulobacter crescentus S-layer protein RsaA N-terminal domain bound to LPS and soaked with Holmium | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, CALCIUM ION, HOLMIUM ATOM, ... | | Authors: | von Kugelgen, A, Bharat, T.A.M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.37 Å) | | Cite: | High-resolution mapping of metal ions reveals principles of surface layer assembly in Caulobacter crescentus cells.
Structure, 30, 2022
|
|
2IBM
 
 | |
7PTT
 
 | |
7PTU
 
 | |
7PTP
 
 | |
7PTR
 
 | |
5U3X
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 8 | | Descriptor: | 6-[2-({cyclopropyl[4-(pyridin-3-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7PYR
 
 | | Crystal structure of the adenosine A2A receptor (A2A-PSB1-bRIL) in complex with preladenant conjugate PSB-2115 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[2-[2-[2-[2-[2-[4-[4-[2-[7-azanyl-4-(furan-2-yl)-3,5,6,8,10,11-hexazatricyclo[7.3.0.0^{2,6}]dodeca-1(9),2,4,7,11-pentaen-10-yl]ethyl]piperazin-1-yl]phenoxy]ethanoylamino]ethoxy]ethoxy]ethoxy]ethoxy]-~{N}-[5-[2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-1,3-diaza-2$l^{4}-boratricyclo[7.3.0.0^{3,7}]dodeca-4,6,9,11-tetraen-8-yl]pentyl]ethanamide, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Claff, T, Klapschinski, T.A, Tiruttani Subhramanyam, U.K, Vaassen, V.J, Schlegel, J.G, Vielmuth, C, Voss, J.H, Labahn, J, Muller, C.E. | | Deposit date: | 2021-10-11 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Single Stabilizing Point Mutation Enables High-Resolution Co-Crystal Structures of the Adenosine A 2A Receptor with Preladenant Conjugates.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7PX4
 
 | | Crystal structure of the adenosine A2A receptor (A2A-PSB1-bRIL) in complex with preladenant conjugate PSB-2113 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Claff, T, Klapschinski, T.A, Tiruttani Subhramanyam, U.K, Vaassen, V.J, Schlegel, J.G, Vielmuth, C, Voss, J.H, Labahn, J, Muller, C.E. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Single Stabilizing Point Mutation Enables High-Resolution Co-Crystal Structures of the Adenosine A 2A Receptor with Preladenant Conjugates.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
4GMZ
 
 | |
5VAF
 
 | |
2FFU
 
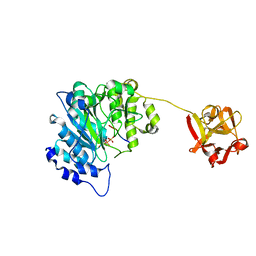 | | Crystal Structure of Human ppGalNAcT-2 complexed with UDP and EA2 | | Descriptor: | 13-Peptide EA2, PTTDSTTPAPTTK, MANGANESE (II) ION, ... | | Authors: | Fritz, T.A. | | Deposit date: | 2005-12-20 | | Release date: | 2006-01-31 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Dynamic Association between the Catalytic and Lectin Domains of Human UDP-GalNAc:Polypeptide {alpha}-N-Acetylgalactosaminyltransferase-2
J.Biol.Chem., 281, 2006
|
|
1TCC
 
 | |
2FE9
 
 | |
5UNH
 
 | | Synchrotron structure of human angiotensin II type 2 receptor in complex with compound 2 (N-[(furan-2-yl)methyl]-N-(4-oxo-2-propyl-3-{[2'-(2H-tetrazol-5-yl)[1,1'- biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)benzamide) | | Descriptor: | N-[(furan-2-yl)methyl]-N-(4-oxo-2-propyl-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)benzamide, Soluble cytochrome b562,Type-2 angiotensin II receptor | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
