2C4I
 
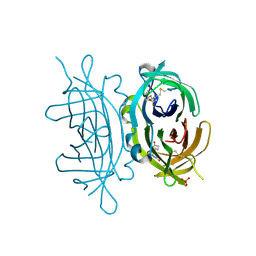 | | Crystal structure of engineered avidin | | Descriptor: | AVIDIN, BIOTIN, SULFATE ION | | Authors: | Hytonen, V.P, Horha, J, Airenne, T.T, Niskanen, E.A, Helttunen, K, Johnson, M.S, Salminen, T.A, Kulomaa, M.S, Nordlund, H.R. | | Deposit date: | 2005-10-19 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Controlling Quaternary Structure Assembly: Subunit Interface Engineering and Crystal Structure of Dual Chain Avidin.
J.Mol.Biol., 359, 2006
|
|
7TPD
 
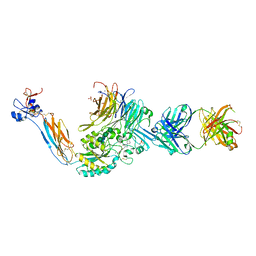 | | Integrin alaphIIBbeta3 complex with EF5154 | | Descriptor: | (4-{[2-oxo-4-(piperidin-4-yl)piperazin-1-yl]acetyl}phenoxy)acetic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhu, J, Lin, F.-Y, Zhu, J, Springer, T.A. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7U9V
 
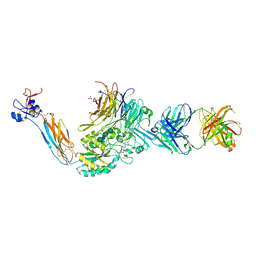 | | Integrin alaphIIBbeta3 complex with BMS4-1 | | Descriptor: | (4-{[(5S)-3-(4-carbamimidoylphenyl)-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperazin-1-yl)acetic acid, 10E5 Fab heavy chain, 10E5 light chain, ... | | Authors: | Zhu, J, Lin, F.-Y, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-11 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25492167 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7THO
 
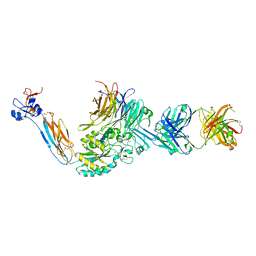 | | Integrin alaphIIBbeta3 complex with Eptifibatide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhu, J, Lin, F.-Y, Zhu, J, Springer, T.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7TMZ
 
 | | Integrin alaphIIBbeta3 complex with BMS compound 4 | | Descriptor: | (4-{[(5S)-3-{4-[(E)-imino(4-methylpiperazin-1-yl)methyl]phenyl}-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperazin-1-yl)acetic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhu, J, Lin, F.-Y, Zhu, J, Springer, T.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.20002 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7U9F
 
 | | Integrin alaphIIBbeta3 complex with BMS compound 4 in Mn2+ | | Descriptor: | (4-{[(5S)-3-{4-[(E)-imino(4-methylpiperazin-1-yl)methyl]phenyl}-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperazin-1-yl)acetic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.70000529 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
7U60
 
 | | Integrin alaphIIBbeta3 complex with cRGDfV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARG-GLY-ASP-DPN-VAL, ... | | Authors: | Zhu, J, Lin, F.Y, Zhu, J, Springer, T.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
2BEJ
 
 | |
3B24
 
 | | Hsp90 alpha N-terminal domain in complex with an aminotriazine fragment molecule | | Descriptor: | 4-(ethylsulfanyl)-6-methyl-1,3,5-triazin-2-amine, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-07-21 | | Release date: | 2011-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lead generation of heat shock protein 90 inhibitors by a combination of fragment-based approach, virtual screening, and structure-based drug design
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7TOD
 
 | | Solution structure of the phosphatidylinositol 3-phosphate binding domain from the Legionella effector SetA | | Descriptor: | Subversion of eukaryotic traffic protein A | | Authors: | Beck, W.H.J, Enoki, T.A, Feigenson, G.W, Nicholson, L, Oswald, R, Mao, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the phosphatidylinositol 3-phosphate binding domain from the Legionella effector SetA
To Be Published
|
|
7TPU
 
 | | Crystal structure of a chitinase-modifying protein from Fusarium vanettenii (Fvan-cmp) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-lactamase domain-containing protein, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Dowling, N.V, Naumann, T.A, Price, N.P.J, Rose, D.R. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of a polyglycine hydrolase determined using a RoseTTAFold model.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3B27
 
 | | Hsp90 alpha N-terminal domain in complex with an inhibitor Ro4919127 | | Descriptor: | 4-(2-chlorophenyl)-6-(methylsulfanyl)-1,3,5-triazin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-07-21 | | Release date: | 2011-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Lead generation of heat shock protein 90 inhibitors by a combination of fragment-based approach, virtual screening, and structure-based drug design
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4JKR
 
 | | Crystal Structure of E. coli RNA Polymerase in complex with ppGpp | | Descriptor: | DNA-DIRECTED RNA POLYMERASE SUBUNIT BETA', DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, Wang, Y, Steitz, T.A. | | Deposit date: | 2013-03-11 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex.
Mol.Cell, 50, 2013
|
|
6ITC
 
 | | Structure of a substrate engaged SecA-SecY protein translocation machine | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ma, C.Y, Wu, X.F, Sun, D.J, Park, E.Y, Rapoport, T.A, Gao, N, Long, L. | | Deposit date: | 2018-11-21 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the substrate-engaged SecA-SecY protein translocation machine.
Nat Commun, 10, 2019
|
|
6HJ4
 
 | | Crystal structure of Whitewater Arroyo virus GP1 glycoprotein at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
8OQ3
 
 | | Structure of methylamine treated human complement C3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gadeberg, T.A.F, Andersen, G.R. | | Deposit date: | 2023-04-11 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of methylamine treated human complement C3
To Be Published
|
|
6HJ5
 
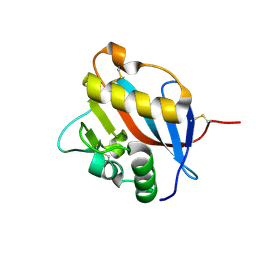 | | Crystal structure of Whitewater Arroyo virus GP1 glycoprotein at pH 5.6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
6HJC
 
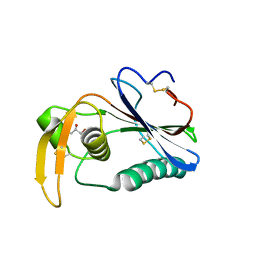 | | Crystal structure of Loei River virus GP1 glycoprotein at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-09-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
7UH4
 
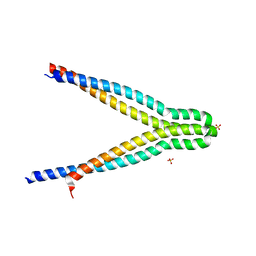 | | LXG-associated alpha-helical protein D2 (LapD2) | | Descriptor: | LXG-associated alpha-helical protein D2, SULFATE ION | | Authors: | Klein, T.A, Grebenc, D.W, Shah, P.Y, McArthur, O.D, Surette, M.G, Kim, Y, Whitney, J.C. | | Deposit date: | 2022-03-25 | | Release date: | 2022-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dual Targeting Factors Are Required for LXG Toxin Export by the Bacterial Type VIIb Secretion System.
Mbio, 13, 2022
|
|
7UBF
 
 | |
7UBD
 
 | |
7UBI
 
 | |
2C1Q
 
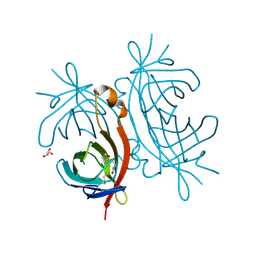 | | X-ray structure of biotin binding protein from chicken | | Descriptor: | BIOTIN, BIOTIN BINDING PROTEIN A, GLYCEROL | | Authors: | Hytonen, V.P, Niskanen, E.A, Maatta, J.A.E, Huuskonen, J, Helttunen, K.J, Halling, K.K, Slotte, J.P, Nordlund, H.R, Rissanen, K, Johnson, M.S, Salminen, T.A, Kulomaa, M.S, Laitinen, O.H, Airenne, T.T. | | Deposit date: | 2005-09-19 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Characterization of a Novel Chicken Biotin-Binding Protein a (Bbp-A).
Bmc Struct.Biol., 7, 2007
|
|
7UZL
 
 | |
7UBC
 
 | |
