3FKD
 
 | |
3EWM
 
 | |
3FCD
 
 | |
3EVZ
 
 | |
3DXI
 
 | |
3DZ1
 
 | |
3DTY
 
 | | Crystal structure of an Oxidoreductase from Pseudomonas syringae | | Descriptor: | MAGNESIUM ION, Oxidoreductase, Gfo/Idh/MocA family | | Authors: | Eswaramoorthy, S, Mahmood, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-16 | | Release date: | 2008-08-05 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of an Oxidoreductase from Pseudomonas syringae
To be Published
|
|
1CT5
 
 | | CRYSTAL STRUCTURE OF YEAST HYPOTHETICAL PROTEIN YBL036C-SELENOMET CRYSTAL | | Descriptor: | PROTEIN (YEAST HYPOTHETICAL PROTEIN, SELENOMET), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-08-18 | | Release date: | 1999-09-02 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a yeast hypothetical protein selected by a structural genomics approach.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3DZB
 
 | |
1CCD
 
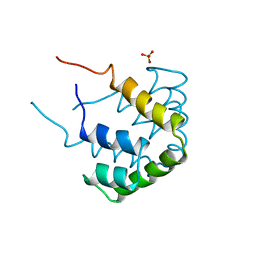 | | REFINED STRUCTURE OF RAT CLARA CELL 17 KDA PROTEIN AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | CLARA CELL 17 kD PROTEIN, SULFATE ION | | Authors: | Umland, T.C, Swaminathan, S, Furey, W, Singh, G, Pletcher, J, Sax, M. | | Deposit date: | 1991-09-17 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Refined structure of rat Clara cell 17 kDa protein at 3.0 A resolution.
J.Mol.Biol., 224, 1992
|
|
3FFZ
 
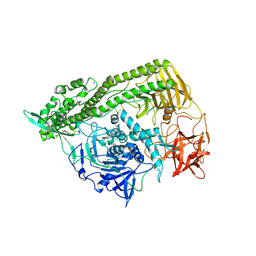 | | Domain organization in Clostridium butulinum neurotoxin type E is unique: Its implication in faster translocation | | Descriptor: | ACETATE ION, Botulinum neurotoxin type E, SODIUM ION, ... | | Authors: | Kumaran, D, Eswaramoorthy, S, Swaminathan, S. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Domain organization in Clostridium botulinum neurotoxin type E is unique: its implication in faster translocation.
J.Mol.Biol., 386, 2009
|
|
3G1W
 
 | |
3EAF
 
 | | Crystal structure of ABC transporter, substrate binding protein Aeropyrum pernix | | Descriptor: | ABC transporter, substrate binding protein, GLYCEROL, ... | | Authors: | Zhang, Z, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-25 | | Release date: | 2008-09-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ABC transporter, substrate binding protein Aeropyrum pernix
To be Published
|
|
3GBU
 
 | | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Uncharacterized sugar kinase PH1459 | | Authors: | Eswaramoorthy, S, Kumar, G, Zhang, Z, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with ATP
To be Published
|
|
3G0O
 
 | |
3G12
 
 | | Crystal structure of a putative lactoylglutathione lyase from Bdellovibrio bacteriovorus | | Descriptor: | Putative lactoylglutathione lyase, SULFATE ION | | Authors: | Patskovsky, Y, Madegowda, M, Gilmore, M, Chang, S, Maletic, M, Smith, D, Sauder, J.M, Burley, S.K, Swaminathan, S, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of a putative lactoylglutathione lyase from Bdellovibrio bacteriovorus
To be Published
|
|
3D0C
 
 | | Crystal structure of dihydrodipicolinate synthase from Oceanobacillus iheyensis at 1.9 A resolution | | Descriptor: | Dihydrodipicolinate synthase | | Authors: | Satyanarayana, L, Eswaramoorthy, S, Sauder, J.M, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-01 | | Release date: | 2008-05-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dihydrodipicolinate synthase from Oceanobacillus iheyensis at 1.9 A resolution.
To be Published
|
|
1F1M
 
 | | CRYSTAL STRUCTURE OF OUTER SURFACE PROTEIN C (OSPC) | | Descriptor: | OUTER SURFACE PROTEIN C, ZINC ION | | Authors: | Kumaran, D, Eswaramoorthy, S, Dunn, J.J, Swaminathan, S. | | Deposit date: | 2000-05-19 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi.
EMBO J., 20, 2001
|
|
3GPK
 
 | |
1F89
 
 | | Crystal structure of Saccharomyces cerevisiae Nit3, a member of branch 10 of the nitrilase superfamily | | Descriptor: | 32.5 KDA PROTEIN YLR351C | | Authors: | Kumaran, D, Eswaramoorthy, S, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-06-29 | | Release date: | 2001-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative CN hydrolase from yeast
Proteins, 52, 2003
|
|
1AF9
 
 | | TETANUS NEUROTOXIN C FRAGMENT | | Descriptor: | TETANUS NEUROTOXIN | | Authors: | Umland, T.C, Wingert, L, Swaminathan, S, Furey, W.F, Schmidt, J.J, Sax, M. | | Deposit date: | 1997-03-24 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the receptor binding fragment HC of tetanus neurotoxin.
Nat.Struct.Biol., 4, 1997
|
|
3EHE
 
 | |
3EEG
 
 | |
3E9M
 
 | |
3EUW
 
 | |
