3HP0
 
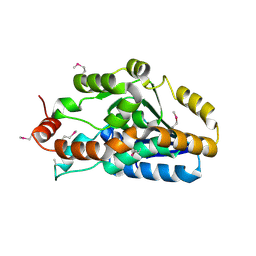 | |
3IH0
 
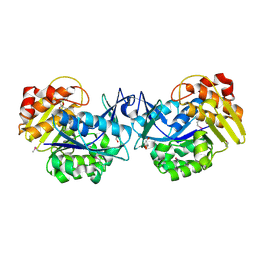 | | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with AMP-PNP | | Descriptor: | GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Uncharacterized sugar kinase PH1459 | | Authors: | Kumar, G, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-29 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with AMP-PNP
To be Published
|
|
1ZL6
 
 | | Crystal structure of Tyr350Ala mutant of Clostridium botulinum neurotoxin E catalytic domain | | Descriptor: | SULFATE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-05 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
1YVG
 
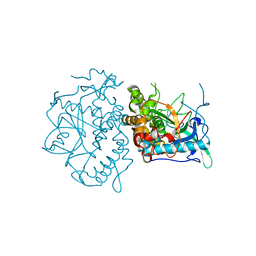 | | Structural analysis of the catalytic domain of tetanus neurotoxin | | Descriptor: | Tetanus toxin, light chain, ZINC ION | | Authors: | Rao, K.N, Kumaran, D, Binz, T, Swaminathan, S. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the catalytic domain of tetanus neurotoxin.
Toxicon, 45, 2005
|
|
1YV9
 
 | |
3MAB
 
 | | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED PROTEIN FROM LISTERIA MONOCYTOGENES, Triclinic FORM | | Descriptor: | uncharacterized protein | | Authors: | Madegowda, M, Chruszcz, M, Minor, W, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of an uncharacterized protein from listeria monocytogenes
To be Published
|
|
3M2T
 
 | |
3OXN
 
 | |
3H49
 
 | |
1Y9Q
 
 | |
1YDF
 
 | |
3G12
 
 | | Crystal structure of a putative lactoylglutathione lyase from Bdellovibrio bacteriovorus | | Descriptor: | Putative lactoylglutathione lyase, SULFATE ION | | Authors: | Patskovsky, Y, Madegowda, M, Gilmore, M, Chang, S, Maletic, M, Smith, D, Sauder, J.M, Burley, S.K, Swaminathan, S, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of a putative lactoylglutathione lyase from Bdellovibrio bacteriovorus
To be Published
|
|
3K2G
 
 | | Crystal structure of a Resiniferatoxin-binding protein from Rhodobacter sphaeroides | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, MAGNESIUM ION, Resiniferatoxin-binding, ... | | Authors: | Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a Resiniferatoxin-binding protein from Rhodobacter sphaeroides
To be Published
|
|
3KXW
 
 | |
3OPN
 
 | |
3U4F
 
 | | Crystal structure of a mandelate racemase (muconate lactonizing enzyme family protein) from Roseovarius nubinhibens | | Descriptor: | GUANIDINE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Eswaramoorthy, S, Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-07 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a mandelate racemase (muconate lactonizing enzyme family protein) from Roseovarius nubinhibens
To be Published, 2011
|
|
3LJS
 
 | |
3E82
 
 | | Crystal structure of a putative oxidoreductase from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, Putative oxidoreductase | | Authors: | Eswaramoorthy, S, Mohammad, M.B, Thomas, C.A, Brown, A.C, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-19 | | Release date: | 2008-08-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of a putative oxidoreductase from Klebsiella pneumoniae
To be Published
|
|
3LDT
 
 | |
3LHL
 
 | | Crystal structure of a putative agmatinase from Clostridium difficile | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Palani, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative agmatinase from Clostridium difficile
To be Published
|
|
3LKI
 
 | |
3EAF
 
 | | Crystal structure of ABC transporter, substrate binding protein Aeropyrum pernix | | Descriptor: | ABC transporter, substrate binding protein, GLYCEROL, ... | | Authors: | Zhang, Z, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-25 | | Release date: | 2008-09-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ABC transporter, substrate binding protein Aeropyrum pernix
To be Published
|
|
1U02
 
 | | Crystal structure of trehalose-6-phosphate phosphatase related protein | | Descriptor: | GLYCEROL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Krishnamurthy, N.R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-12 | | Release date: | 2004-07-20 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of trehalose-6-phosphate phosphatase-related protein: biochemical and biological implications.
Protein Sci., 15, 2006
|
|
1TXN
 
 | |
3LUA
 
 | |
