7W05
 
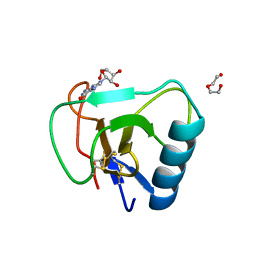 | | 12 mutant Ribonuclease from Hericium erinaceus GMP binding form | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE, Ribonuclease T1 | | Authors: | Takebe, K, Chida, T, Suzuki, M, Itagaki, T, Morita, Y, Uzawa, N, Kobayashi, H. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | 12 mutant Ribonuclease from Hericium erinaceus GMP binding form
To Be Published
|
|
2DCZ
 
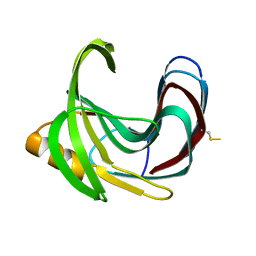 | | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase By Directed Evolution | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Endo-1,4-beta-xylanase A, SULFATE ION | | Authors: | Kondo, H, Miyazaki, K, Takenouchi, M, Noro, N, Suzuki, M, Tsuda, S. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase by Directed Evolution
J.Biol.Chem., 281, 2006
|
|
2DCY
 
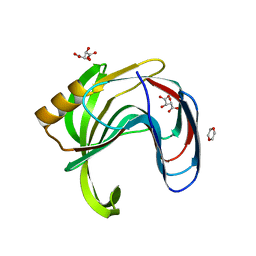 | | Crystal structure of Bacillus subtilis family-11 xylanase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, D(-)-TARTARIC ACID, Endo-1,4-beta-xylanase A, ... | | Authors: | Kondo, H, Miyazaki, K, Takenouchi, M, Noro, N, Suzuki, M, Tsuda, S. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase by Directed Evolution
J.Biol.Chem., 281, 2006
|
|
2E1C
 
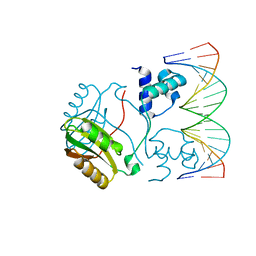 | | Structure of Putative HTH-type transcriptional regulator PH1519/DNA Complex | | Descriptor: | DNA (5'-D(*DAP*DGP*DTP*DGP*DAP*DAP*DAP*DAP*DTP*DTP*DTP*DTP*DTP*DCP*DAP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DTP*DGP*DAP*DAP*DAP*DAP*DAP*DTP*DTP*DTP*DTP*DCP*DAP*DCP*DT)-3'), Putative HTH-type transcriptional regulator PH1519 | | Authors: | Koike, H, Suzuki, M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-12-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Feast/Famine Regulation by Transcription Factor FL11 for the Survival of the Hyperthermophilic Archaeon Pyrococcus OT3.
Structure, 15, 2007
|
|
2E5A
 
 | | Crystal Structure of Bovine Lipoyltransferase in Complex with Lipoyl-AMP | | Descriptor: | 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, ACETIC ACID, Lipoyltransferase 1, ... | | Authors: | Fujiwara, K, Hosaka, H, Matsuda, M, Suzuki, M, Nakagawa, A. | | Deposit date: | 2006-12-19 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of bovine Lipoyltransferase in complex with lipoyl-AMP
J.Mol.Biol., 371, 2007
|
|
1IU1
 
 | | Crystal structure of human gamma1-adaptin ear domain | | Descriptor: | gamma1-adaptin | | Authors: | Nogi, T, Shiba, Y, Kawasaki, M, Shiba, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Takatsu, H, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2002-02-19 | | Release date: | 2002-07-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the accessory protein recruitment by the gamma-adaptin ear domain.
Nat.Struct.Biol., 9, 2002
|
|
1J2J
 
 | | Crystal structure of GGA1 GAT N-terminal region in complex with ARF1 GTP form | | Descriptor: | ADP-ribosylation factor 1, ADP-ribosylation factor binding protein GGA1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-01-05 | | Release date: | 2003-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport
NAT.STRUCT.BIOL., 10, 2003
|
|
1JWG
 
 | | VHS Domain of human GGA1 complexed with cation-independent M6PR C-terminal Peptide | | Descriptor: | ADP-ribosylation factor binding protein GGA1, Cation-independent mannose-6-phosphate receptor, IODIDE ION | | Authors: | Shiba, T, Takatsu, H, Nogi, T, Matsugaki, N, Kawasaki, M, Igarashi, N, Suzuki, M, Kato, R, Earnest, T, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2001-09-04 | | Release date: | 2002-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for recognition of acidic-cluster dileucine sequence by GGA1.
Nature, 415, 2002
|
|
1JWF
 
 | | Crystal Structure of human GGA1 VHS domain. | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Shiba, T, Takatsu, H, Nogi, T, Matsugaki, N, Kawasaki, M, Igarashi, N, Suzuki, M, Kato, R, Earnest, T, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2001-09-04 | | Release date: | 2002-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of acidic-cluster dileucine sequence by GGA1.
Nature, 415, 2002
|
|
1HI2
 
 | | Eosinophil-derived Neurotoxin (EDN) - Sulphate Complex | | Descriptor: | EOSINOPHIL-DERIVED NEUROTOXIN, SULFATE ION | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI5
 
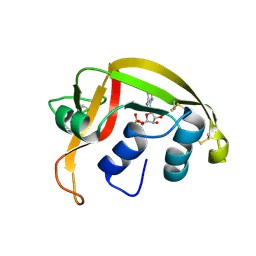 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI4
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosien-3'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
2D05
 
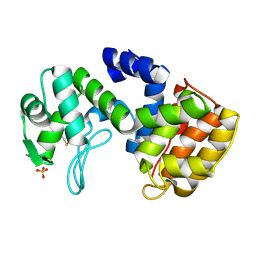 | | Chitosanase From Bacillus circulans mutant K218P | | Descriptor: | Chitosanase, SULFATE ION | | Authors: | Fukamizo, T, Amano, S, Yamaguchi, K, Yoshikawa, T, Katsumi, T, Saito, J, Suzuki, M, Miki, K, Nagata, Y, Ando, A. | | Deposit date: | 2005-07-25 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus circulans MH-K1 Chitosanase: Amino Acid Residues Responsible for Substrate Binding
J.Biochem.(Tokyo), 138, 2005
|
|
5GY6
 
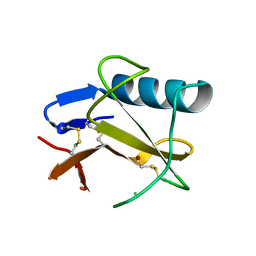 | | Ribonuclease from Hericium erinaceus (RNase He1) | | Descriptor: | Ribonuclease T1, ZINC ION | | Authors: | Kobayashi, H, Sangawa, T, Takebe, K, Itagaki, T, Motoyoshi, N, Suzuki, M. | | Deposit date: | 2016-09-21 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ribonuclease from Hericium erinaceus (RNase He1)
To Be Published
|
|
2E0Z
 
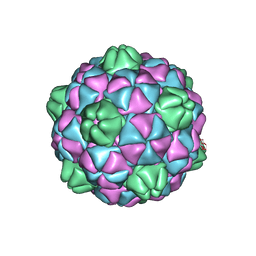 | | Crystal structure of virus-like particle from Pyrococcus furiosus | | Descriptor: | Virus-like particle | | Authors: | Akita, F, Chong, K.T, Tanaka, H, Yamashita, E, Miyazaki, N, Nakaishi, Y, Namba, K, Ono, Y, Suzuki, M, Tsukihara, T, Nakagawa, A. | | Deposit date: | 2006-10-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Crystal Structure of a Virus-like Particle from the Hyperthermophilic Archaeon Pyrococcus furiosus Provides Insight into the Evolution of Viruses
J.Mol.Biol., 368, 2007
|
|
5GQP
 
 | | Thaumatin Structure at pH 8.0, orthorhombic type1 | | Descriptor: | Thaumatin I | | Authors: | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | Thaumatin Structure at pH 8.0, orthorhombic type1
To Be Published
|
|
5Y5N
 
 | | Crystal structure of human Sirtuin 2 in complex with a selective inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]benzamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Mellini, P, Itoh, Y, Tsumoto, H, Li, Y, Suzuki, M, Tokuda, N, Kakizawa, T, Miura, Y, Takeuchi, J, Lahtela-Kakkonen, M, Suzuki, T. | | Deposit date: | 2017-08-09 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent mechanism-based sirtuin-2-selective inhibition by anin situ-generated occupant of the substrate-binding site, "selectivity pocket" and NAD+-binding site.
Chem Sci, 8, 2017
|
|
4YSD
 
 | | Room temperature structure of copper nitrite reductase from Geobacillus thermodenitrificans | | Descriptor: | ACETIC ACID, COPPER (II) ION, Nitrite reductase | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSQ
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 8.38 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSP
 
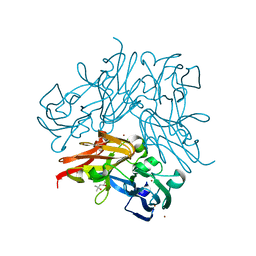 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 8.32 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diedrichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
1ISP
 
 | | Crystal structure of Bacillus subtilis lipase at 1.3A resolution | | Descriptor: | GLYCEROL, lipase | | Authors: | Kawasaki, K, Kondo, H, Suzuki, M, Ohgiya, S, Tsuda, S. | | Deposit date: | 2001-12-19 | | Release date: | 2002-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Alternate conformations observed in catalytic serine of Bacillus subtilis lipase determined at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4YSC
 
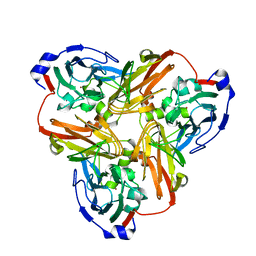 | | Completely oxidized structure of copper nitrite reductase from Alcaligenes faecalis | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YSR
 
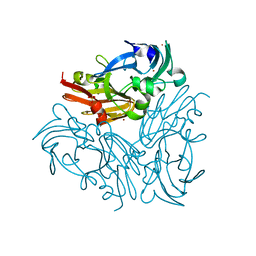 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 16.6 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSS
 
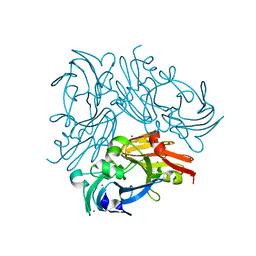 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 16.7 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diedrichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSO
 
 | | Copper nitrite reductase from Geobacillus thermodenitrificans - 0.064 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
