3AD7
 
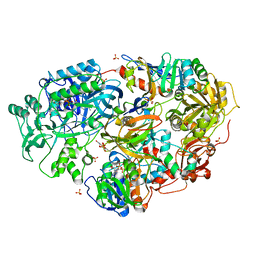 | | Heterotetrameric Sarcosine Oxidase from Corynebacterium sp. U-96 in complex with methylthio acetate | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, H, Moriguchi, T, Ida, K. | | Deposit date: | 2010-01-15 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Channeling and conformational changes in the heterotetrameric sarcosine oxidase from Corynebacterium sp. U-96.
J.Biochem., 148, 2010
|
|
3AD9
 
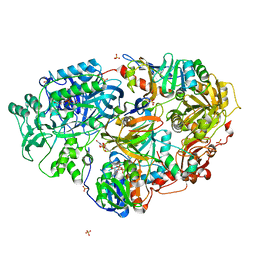 | | Heterotetrameric Sarcosine Oxidase from Corynebacterium sp. U-96 sarcosine-reduced form | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, H, Moriguchi, T, Ida, K. | | Deposit date: | 2010-01-15 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Channeling and conformational changes in the heterotetrameric sarcosine oxidase from Corynebacterium sp. U-96.
J.Biochem., 148, 2010
|
|
3AYL
 
 | |
3ADA
 
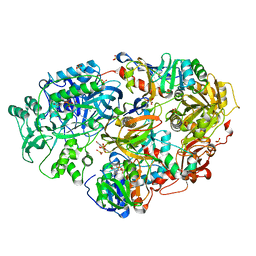 | | Heterotetrameric Sarcosine Oxidase from Corynebacterium sp. U-96 in complex with sulfite | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, H, Moriguchi, T, Ida, K. | | Deposit date: | 2010-01-15 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Channeling and conformational changes in the heterotetrameric sarcosine oxidase from Corynebacterium sp. U-96.
J.Biochem., 148, 2010
|
|
3VTV
 
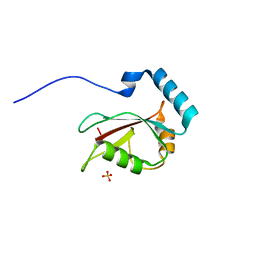 | | Crystal structure of Optineurin LIR-fused human LC3B_2-119 | | Descriptor: | Optineurin, microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3WAL
 
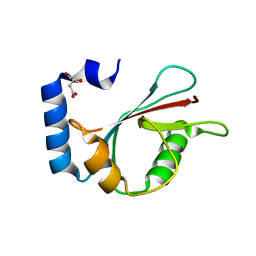 | | Crystal structure of human LC3A_2-121 | | Descriptor: | D-MALATE, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3VTU
 
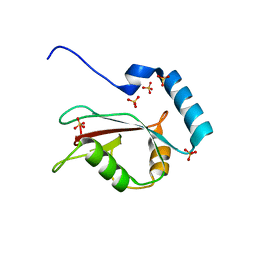 | | Crystal structure of human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3VTW
 
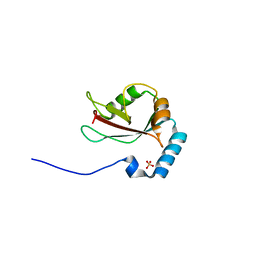 | | Crystal structure of T7-tagged Optineurin LIR-fused human LC3B_2-119 | | Descriptor: | Optineurin, microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3WAO
 
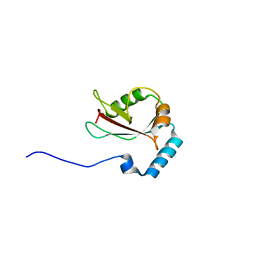 | | Crystal structure of Atg13 LIR-fused human LC3B_2-119 | | Descriptor: | Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAN
 
 | | Crystal structure of Atg13 LIR-fused human LC3A_2-121 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAM
 
 | | Crystal structure of human LC3C_8-125 | | Descriptor: | CITRIC ACID, Microtubule-associated proteins 1A/1B light chain 3C | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAP
 
 | | Crystal structure of Atg13 LIR-fused human LC3C_8-125 | | Descriptor: | Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3C | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3X0W
 
 | | Crystal structure of PLEKHM1 LIR-fused human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, McEwan, D.G, Popovic, D, Gubas, A, Terawaki, S, Stadel, D, Coxon, F, Stegmann, D.M, Bhogaraju, S, Maddi, K, Kirchhoff, A, Gatti, E, Helfrich, M.H, Behrends, C, Pierre, P, Dikic, I, Wakatsuki, S. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins.
Mol.Cell, 57, 2015
|
|
4TLJ
 
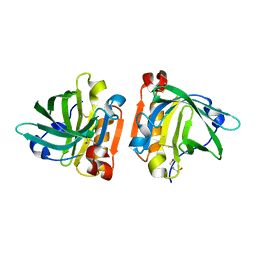 | | Ultra-high resolution crystal structure of caprine Beta-lactoglobulin | | Descriptor: | 1,4-BUTANEDIOL, Beta-lactoglobulin | | Authors: | Crowther, J.M, Jameson, G.B, Suzuki, H, Dobson, R.C.J. | | Deposit date: | 2014-05-30 | | Release date: | 2014-06-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Ultra-high resolution crystal structure of recombinant caprine beta-lactoglobulin.
Febs Lett., 588, 2014
|
|
6ORB
 
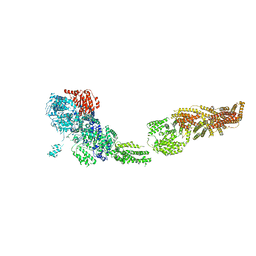 | | Full-length S. pombe Mdn1 in the presence of ATP and Rbin-1 | | Descriptor: | Midasin | | Authors: | Chen, Z, Suzuki, H, Wang, A.C, DiMaio, F, Walz, T, Kapoor, T.M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Cell, 175, 2018
|
|
6OR5
 
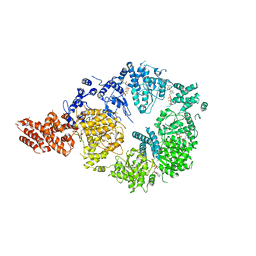 | | Full-length S. pombe Mdn1 in the presence of AMPPNP (ring region) | | Descriptor: | Midasin, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Chen, Z, Suzuki, H, Wang, A.C, DiMaio, F, Walz, T, Kapoor, T.M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Cell, 175, 2018
|
|
3IYZ
 
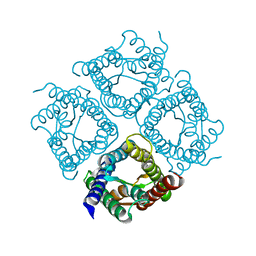 | | Structure of Aquaporin-4 S180D mutant at 10.0 A resolution from electron micrograph | | Descriptor: | Aquaporin-4 | | Authors: | Mitsuma, T, Tani, K, Hiroaki, Y, Kamegawa, A, Suzuki, H, Hibino, H, Kurachi, Y, Fujiyoshi, Y. | | Deposit date: | 2010-07-24 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Influence of the cytoplasmic domains of aquaporin-4 on water conduction and array formation.
J.Mol.Biol., 402, 2010
|
|
8GCL
 
 | | Cryo-EM structure of hAQP2 in DDM | | Descriptor: | Aquaporin-2 | | Authors: | Kamegawa, A, Suzuki, S, Nishikawa, K, Numoto, N, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-03-02 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural analysis of the water channel AQP2 by single-particle cryo-EM.
J.Struct.Biol., 215, 2023
|
|
8IHI
 
 | | Cryo-EM structure of HCA2-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHF
 
 | | Cryo-EM structure of HCA2-Gi complex with MK6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHK
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran (local) | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Soluble cytochrome b562,Hydroxycarboxylic acid receptor 3 | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHJ
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHH
 
 | | Cryo-EM structure of HCA2-Gi complex with LUF6283 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-butyl-1~{H}-pyrazole-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHB
 
 | | Cryo-EM structure of HCA2-Gi complex with GSK256073 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
6OR6
 
 | | Full-length S. pombe Mdn1 in the presence of AMPPNP (tail region) | | Descriptor: | Midasin | | Authors: | Chen, Z, Suzuki, H, Wang, A.C, DiMaio, F, Walz, T, Kapoor, T.M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Cell, 175, 2018
|
|
