1XBH
 
 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(L-262) | | Descriptor: | PROTEIN (CYCLO(L-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-02-17 | | Release date: | 1999-02-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
6EYO
 
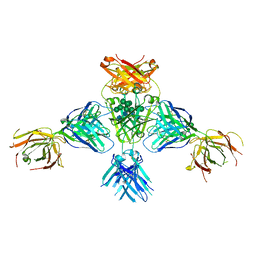 | | Structure of extended IgE-Fc in complex with two anti-IgE Fabs | | Descriptor: | 8D6 Fab heavy chain, 8D6 Fab light chain, Immunoglobulin heavy constant epsilon, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
6EYN
 
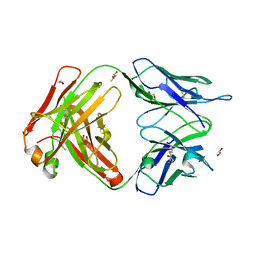 | | Structure of the 8D6 (anti-IgE) Fab | | Descriptor: | 1,2-ETHANEDIOL, 8D6 Fab heavy chain, 8D6 Fab light chain, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
3FCZ
 
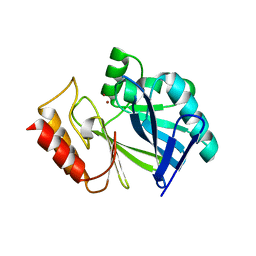 | | Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility | | Descriptor: | Beta-lactamase 2, ZINC ION | | Authors: | Tomatis, P, Fabiane, S, Simona, F, Carloni, P, Sutton, B, Vila, A. | | Deposit date: | 2008-11-24 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6HRJ
 
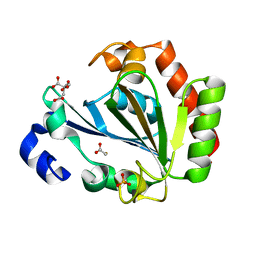 | | Apo - YndL | | Descriptor: | CITRATE ANION, SULFATE ION, YndL, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
FEBS J., 285, 2018
|
|
6HRI
 
 | | Native YndL | | Descriptor: | CITRATE ANION, IMIDAZOLE, SULFATE ION, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
FEBS J., 285, 2018
|
|
1C4B
 
 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(RD-262) | | Descriptor: | PROTEIN (CYCLO(RD-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-08-02 | | Release date: | 1999-08-25 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
1ADQ
 
 | |
1G84
 
 | | THE SOLUTION STRUCTURE OF THE C EPSILON2 DOMAIN FROM IGE | | Descriptor: | IMMUNOGLOBULIN E | | Authors: | McDonnell, J.M, Cowburn, D, Gould, H.J, Sutton, B.J, Calvert, R, Beavil, R.E, Beavil, A.J, Henry, A.J. | | Deposit date: | 2000-11-16 | | Release date: | 2001-05-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structure of the IgE Cepsilon2 domain and its role in stabilizing the complex with its high-affinity receptor FcepsilonRIalpha.
Nat.Struct.Biol., 8, 2001
|
|
1QOK
 
 | | MFE-23 AN ANTI-CARCINOEMBRYONIC ANTIGEN SINGLE-CHAIN FV ANTIBODY | | Descriptor: | MFE-23 RECOMBINANT ANTIBODY FRAGMENT | | Authors: | Boehm, M.K, Corper, A.L, Wan, T, Sohi, M.K, Sutton, B.J, Thornton, J.D, Keep, P.A, Chester, K.A, Begent, R.H.J, Perkins, S.J. | | Deposit date: | 1999-11-11 | | Release date: | 2000-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Anti-Carcinoembryonic Antigen Single-Chain Fv Antibody Mfe-23 and a Model for Antigen Binding Based on Intermolecular Contacts
Biochem.J., 346, 2000
|
|
6Y0L
 
 | | Crystal structure of human CD23 lectin domain N225D, K229E, S252N, T251N, R253G, S254G mutant | | Descriptor: | GLYCEROL, Low affinity immunoglobulin epsilon Fc receptor membrane-bound form, SULFATE ION | | Authors: | Ilkow, V.F, Davies, A.M, Sutton, B.J, McDonnell, J.M. | | Deposit date: | 2020-02-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Reviving lost binding sites: Exploring calcium-binding site transitions between human and murine CD23.
Febs Open Bio, 11, 2021
|
|
6Y0M
 
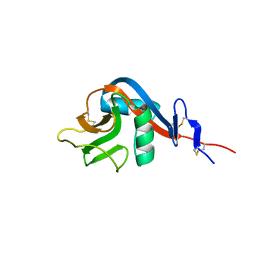 | | Crystal structure of human CD23 lectin domain N225D, K229E, S252N, T251N mutant | | Descriptor: | Low affinity immunoglobulin epsilon Fc receptor membrane-bound form | | Authors: | Ilkow, V.F, Davies, A.M, Sutton, B.J, McDonnell, J.M. | | Deposit date: | 2020-02-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reviving lost binding sites: Exploring calcium-binding site transitions between human and murine CD23.
Febs Open Bio, 11, 2021
|
|
5G64
 
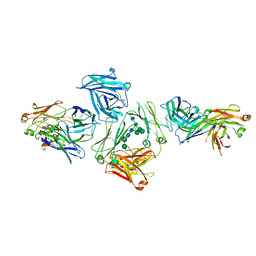 | | The complex between human IgE-Fc and two anti-IgE Fab fragments | | Descriptor: | FAB FRAGMENT, IG EPSILON CHAIN C REGION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Davies, A.M, Allan, E.G, Keeble, A.H, Delgado, J, Cossins, B.P, Mitropoulou, A.N, Pang, M.O.Y, Ceska, T, Beavil, A.J, Craggs, G, Westwood, M, Henry, A.J, McDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-06-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.715 Å) | | Cite: | Allosteric mechanism of action of the therapeutic anti-IgE antibody omalizumab.
J. Biol. Chem., 292, 2017
|
|
1HEZ
 
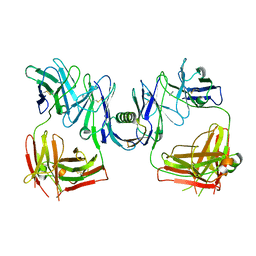 | | Structure of P. magnus protein L bound to a human IgM Fab. | | Descriptor: | HEAVY CHAIN OF IG, IMIDAZOLE, KAPPA LIGHT CHAIN OF IG, ... | | Authors: | Graille, M, Stura, E.A, Housden, N.G, Bottomley, S.P, Taussig, M.J, Sutton, B.J, Gore, M.G, Charbonnier, J.B. | | Deposit date: | 2000-11-27 | | Release date: | 2001-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex between Peptostreptococcus Magnus Protein L and a Human Antibody Reveals Structural Convergence in the Interaction Modes of Fab Binding Modes
Structure, 9, 2001
|
|
1T8C
 
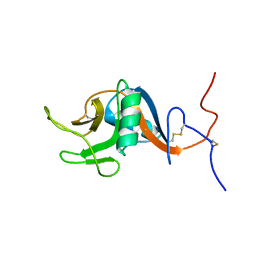 | | Structure of the C-type lectin domain of CD23 | | Descriptor: | Low affinity immunoglobulin epsilon Fc receptor | | Authors: | Hibbert, R.G, Teriete, P, Grundy, G.J, Sutton, B.J, Gould, H.J, McDonnell, J.M. | | Deposit date: | 2004-05-12 | | Release date: | 2005-07-26 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The structure of human CD23 and its interactions with IgE and CD21
J.Exp.Med., 202, 2005
|
|
5ONJ
 
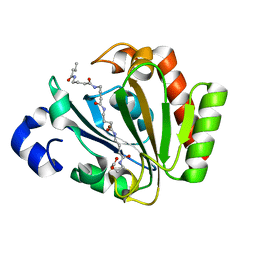 | | YnDL in Complex with 5 amino acid (PGA) complex | | Descriptor: | (2R,3S,4R,5R,6R)-6-((1R,2R,3S,4R,6S)-4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYLOXY)-5-AMINO-2-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, YndL, ZINC ION | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
Febs J., 285, 2018
|
|
1T8D
 
 | | Structure of the C-type lectin domain of CD23 | | Descriptor: | Low affinity immunoglobulin epsilon Fc receptor | | Authors: | Hibbert, R.G, Teriete, P, Grundy, G.J, Sutton, B.J, Gould, H.J, McDonnell, J.M. | | Deposit date: | 2004-05-12 | | Release date: | 2005-07-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structure of human CD23 and its interactions with IgE and CD21
J.Exp.Med., 202, 2005
|
|
2J8R
 
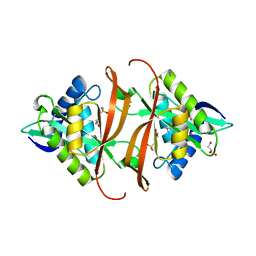 | | Structure of P. aeruginosa acetyltransferase PA4866 solved in complex with L-Methionine sulfoximine | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, ACETYLTRANSFERASE PA4866 FROM P. AERUGINOSA, AZIDE ION, ... | | Authors: | Davies, A.M, Tata, R, Beavil, R.L, Sutton, B.J, Brown, P.R. | | Deposit date: | 2006-10-27 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | L-Methionine Sulfoximine, But not Phosphinothricin, is a Substrate for an Acetyltransferase (Gene Pa4866) from Pseudomonas Aeruginosa: Structural and Functional Studies.
Biochemistry, 46, 2007
|
|
2J8N
 
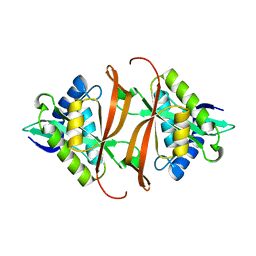 | | Structure of P. aeruginosa acetyltransferase PA4866 solved at room temperature | | Descriptor: | ACETYLTRANSFERASE PA4866 FROM P. AERUGINOSA | | Authors: | Davies, A.M, Tata, R, Beavil, R.L, Sutton, B.J, Brown, P.R. | | Deposit date: | 2006-10-26 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | l-Methionine sulfoximine, but not phosphinothricin, is a substrate for an acetyltransferase (gene PA4866) from Pseudomonas aeruginosa: structural and functional studies.
Biochemistry, 46, 2007
|
|
1BC2
 
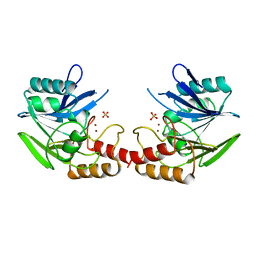 | | ZN-DEPENDENT METALLO-BETA-LACTAMASE FROM BACILLUS CEREUS | | Descriptor: | METALLO-BETA-LACTAMASE II, SULFATE ION, ZINC ION | | Authors: | Fabiane, S.M, Sutton, B.J. | | Deposit date: | 1997-04-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the zinc-dependent beta-lactamase from Bacillus cereus at 1.9 A resolution: binuclear active site with features of a mononuclear enzyme.
Biochemistry, 37, 1998
|
|
1O0V
 
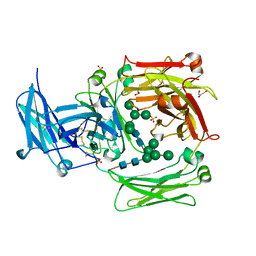 | | The crystal structure of IgE Fc reveals an asymmetrically bent conformation | | Descriptor: | GLYCEROL, Immunoglobulin heavy chain epsilon-1, SULFATE ION, ... | | Authors: | Wan, T, Beavil, R.L, Fabiane, S.M, Beavil, A.J, Sohi, M.K, Keown, M, Young, R.J, Henry, A.J, Owens, R.J, Gould, H.J, Sutton, B.J. | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of IgE Fc reveals an asymmetrically bent conformation
NAT.IMMUNOL., 3, 2002
|
|
6TCR
 
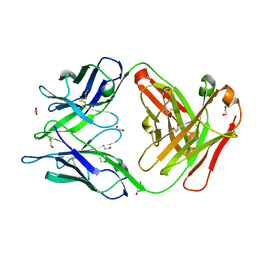 | | Crystal structure of the omalizumab Fab Ser81Arg, Gln83Arg and Leu158Pro light chain mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Omalizumab Fab Ser81Arg, ... | | Authors: | Mitropoulou, A.N, Ceska, T, Beavil, A.J, Henry, A.J, McDonnell, J.M, Sutton, B.J, Davies, A.M. | | Deposit date: | 2019-11-06 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Engineering the Fab fragment of the anti-IgE omalizumab to prevent Fab crystallization and permit IgE-Fc complex crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TCM
 
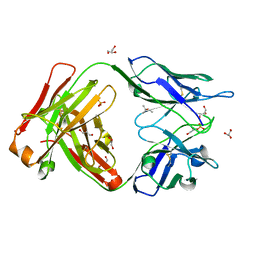 | | Crystal structure of the omalizumab Fab - crystal form I | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Omalizumab Fab, ... | | Authors: | Mitropoulou, A.N, Ceska, T, Beavil, A.J, Henry, A.J, McDonnell, J.M, Sutton, B.J, Davies, A.M. | | Deposit date: | 2019-11-06 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering the Fab fragment of the anti-IgE omalizumab to prevent Fab crystallization and permit IgE-Fc complex crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TCS
 
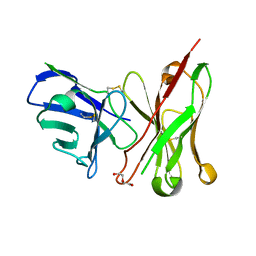 | | Crystal structure of the omalizumab scFv | | Descriptor: | DI(HYDROXYETHYL)ETHER, Omalizumab scFv | | Authors: | Mitropoulou, A.N, Ceska, T, Beavil, A.J, Henry, A.J, McDonnell, J.M, Sutton, B.J, Davies, A.M. | | Deposit date: | 2019-11-06 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering the Fab fragment of the anti-IgE omalizumab to prevent Fab crystallization and permit IgE-Fc complex crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5ONK
 
 | | Native YndL | | Descriptor: | CITRATE ANION, IMIDAZOLE, SULFATE ION, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
FEBS J., 285, 2018
|
|
