3TEE
 
 | | Crystal Structure of Salmonella FlgA in open form | | Descriptor: | CHLORIDE ION, Flagella basal body P-ring formation protein flgA, GLYCEROL | | Authors: | Matsunami, H, Samatey, F.A, Namba, K. | | Deposit date: | 2011-08-12 | | Release date: | 2012-08-15 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural flexibility of the periplasmic protein, FlgA, regulates flagellar P-ring assembly in Salmonella enterica
Sci Rep, 6, 2016
|
|
3VJP
 
 | |
2EJY
 
 | |
2EV8
 
 | |
4QY1
 
 | | Structure of H10 from human-infecting H10N8 in complex with avian receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4X1D
 
 | | Ytterbium-bound human serum transferrin | | Descriptor: | GLYCEROL, MALONATE ION, Serotransferrin, ... | | Authors: | Wang, M, Zhang, H, Sun, H. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | "Anion clamp" allows flexible protein to impose coordination geometry on metal ions
Chem.Commun.(Camb.), 51, 2015
|
|
4X1B
 
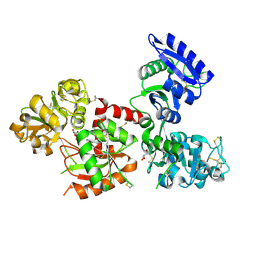 | | Human serum transferrin with ferric ion bound at the C-lobe only | | Descriptor: | FE (III) ION, GLYCEROL, MALONATE ION, ... | | Authors: | Wang, M, Zhang, H, Sun, H. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | "Anion clamp" allows flexible protein to impose coordination geometry on metal ions
Chem.Commun.(Camb.), 51, 2015
|
|
4TQK
 
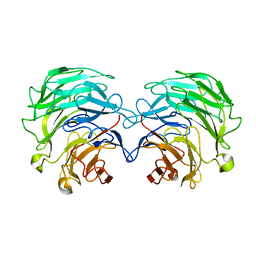 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lectin 2 | | Authors: | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
4TQM
 
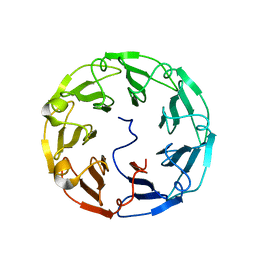 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lectin 2 | | Authors: | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
4TQJ
 
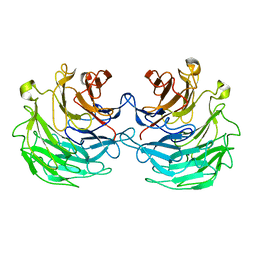 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | Descriptor: | Lectin 2 | | Authors: | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
6LRL
 
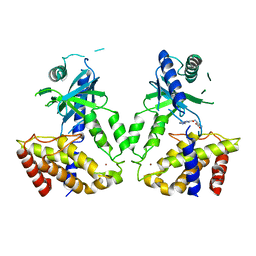 | | Human cGAS catalytic domain bound with compound s2 | | Descriptor: | 3-[[5-(1,2,4-triazol-4-yl)-4H-1,2,4-triazol-3-yl]carbonylamino]benzoic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
6LRJ
 
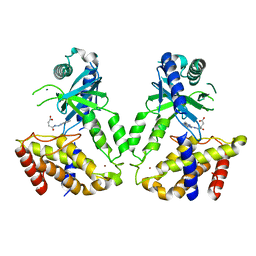 | | Human cGAS catalytic domain bound with compound 23 | | Descriptor: | 4-[2-(2-methyl-[1,2,4]triazolo[1,5-c]quinazolin-5-yl)hydrazinyl]-4-oxidanylidene-butanoic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
6LRI
 
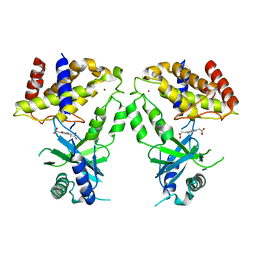 | | Human cGAS catalytic domain bound with compound 17 | | Descriptor: | 3-[5-(2-hydroxy-2-oxoethyl)-3-oxidanylidene-[1,2,4]triazino[2,3-a]benzimidazol-2-yl]propanoic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
6LSA
 
 | | Complex structure of bovine herpesvirus 1 glycoprotein D and bovine nectin-1 IgV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D, ... | | Authors: | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
6LRE
 
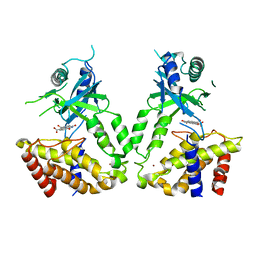 | | Human cGAS catalytic domain bound with compound 3 | | Descriptor: | 1,3-bis(oxidanylidene)benzo[de]isoquinoline-6,7-dicarboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
6LRK
 
 | | Human cGAS catalytic domain bound with compound 40 | | Descriptor: | (3R)-1-pyrrolo[1,2-a]quinoxalin-4-ylpiperidine-3-carboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Zhao, W.F, Xiong, M.Y, Yuan, X.J, Sun, H.B, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
6LS9
 
 | | Crystal structure of bovine herpesvirus 1 glycoprotein D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D | | Authors: | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
9IUM
 
 | |
9IUK
 
 | | The structure of Candida albicans Cdr1 in apo state | | Descriptor: | Pip2(20:4/18:0), Pleiotropic ABC efflux transporter of multiple drugs CDR1 | | Authors: | Peng, Y, Sun, H, Yan, Z.F. | | Deposit date: | 2024-07-22 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of Candida albicans Cdr1 reveal azole-substrate recognition and inhibitor blocking mechanisms.
Nat Commun, 15, 2024
|
|
9IUL
 
 | | The structure of Candida albicans Cdr1 in fluconazole-bound state | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Pip2(20:4/18:0), Pleiotropic ABC efflux transporter of multiple drugs CDR1 | | Authors: | Peng, Y, Sun, H, Yan, Z.F. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of Candida albicans Cdr1 reveal azole-substrate recognition and inhibitor blocking mechanisms.
Nat Commun, 15, 2024
|
|
7X3Y
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (CVB1-E:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-01 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X35
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3M
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07045 | | Descriptor: | (2~{R})-2-[4-[3,5-bis(chloranyl)phenyl]-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X3L
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07044 | | Descriptor: | (2~{R})-2-[4-(3-fluoranyl-4-methyl-phenyl)-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
6LI5
 
 | | Crystal structure of apo-MCR-1-S | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1 | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
