4YSU
 
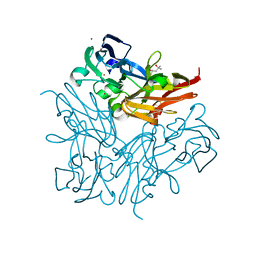 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 25.0 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
7VJ7
 
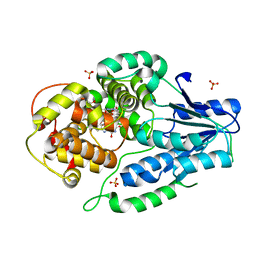 | | SFX structure of archaeal class II CPD photolyase from Methanosarcina mazei in the fully reduced state | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
4YSE
 
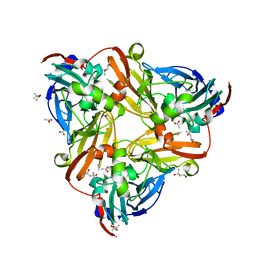 | | High resolution synchrotron structure of copper nitrite reductase from Alcaligenes faecalis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1Y43
 
 | | crystal structure of aspergilloglutamic peptidase from Aspergillus niger | | Descriptor: | Aspergillopepsin II heavy chain, Aspergillopepsin II light chain, SULFATE ION | | Authors: | Sasaki, H, Nakagawa, A, Iwata, S, Muramatsu, T, Suganuma, M, Sawano, Y, Kojima, M, Kubota, K, Takahashi, K. | | Deposit date: | 2004-11-30 | | Release date: | 2005-12-13 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The three-dimensional structure of aspergilloglutamic peptidase from Aspergillus niger
Proc.Jpn.Acad.,Ser.B, 80, 2004
|
|
1V3V
 
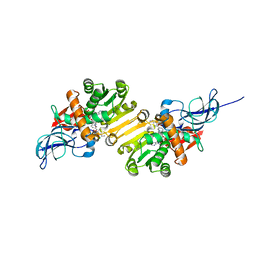 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complexed with NADP and 15-oxo-PGE2 | | Descriptor: | (5E,13E)-11-HYDROXY-9,15-DIOXOPROSTA-5,13-DIEN-1-OIC ACID, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-06 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
1V3T
 
 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
1V26
 
 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MYRISTIC ACID, ... | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-07 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of the Substrate-specific Two-step Catalysis of Long Chain Fatty Acyl-CoA Synthetase Dimer
J.Biol.Chem., 279, 2004
|
|
5X9M
 
 | | Structure of hyper-sweet thaumatin (D21N) | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Masuda, T, Okubo, K, Sugahara, M, Suzuki, M, Mikami, B. | | Deposit date: | 2017-03-08 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Subatomic structure of hyper-sweet thaumatin D21N mutant reveals the importance of flexible conformations for enhanced sweetness.
Biochimie, 157, 2019
|
|
1ULU
 
 | | Crystal structure of tt0143 from Thermus thermophilus HB8 | | Descriptor: | enoyl-acyl carrier protein reductase | | Authors: | Ago, H, Hamada, K, Ida, K, Kanda, H, Sugahara, M, Yamamoto, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of tt0143 from Thermus thermophilus HB8
To be Published
|
|
1ULT
 
 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | CITRIC ACID, long chain fatty acid-CoA ligase | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of the substrate specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer
J.Biol.Chem., 279, 2004
|
|
5X9L
 
 | | Recombinant thaumatin I at 0.9 Angstrom | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Masuda, T, Okubo, K, Sugahara, M, Suzuki, M, Mikami, B. | | Deposit date: | 2017-03-08 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Subatomic structure of hyper-sweet thaumatin D21N mutant reveals the importance of flexible conformations for enhanced sweetness.
Biochimie, 157, 2019
|
|
1ULQ
 
 | | Crystal structure of tt0182 from Thermus thermophilus HB8 | | Descriptor: | putative acetyl-CoA acetyltransferase | | Authors: | Ago, H, Hamada, K, Ida, K, Kanda, H, Sugahara, M, Yamamoto, M, Kuroishi, C, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of tt0182 from Thermus thermophilus HB8
To be Published
|
|
1V25
 
 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, long-chain-fatty-acid-CoA synthetase | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-07 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Substrate-specific Two-step Catalysis of Long Chain Fatty Acyl-CoA Synthetase Dimer
J.Biol.Chem., 279, 2004
|
|
1V3U
 
 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase in apo form | | Descriptor: | CHLORIDE ION, leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
1TUO
 
 | | Crystal structure of putative phosphomannomutase from Thermus Thermophilus HB8 | | Descriptor: | Putative phosphomannomutase | | Authors: | Misaki, S, Suzuki, S, Fujimoto, S, Sakurai, M, Kobayashi, M, Nishijima, K, Kunishima, N, Sugawara, M, Kuroishi, C, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-25 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative phosphomannomutase from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
2CWE
 
 | | Crystal structure of hypothetical transcriptional regulator protein, PH1932 from Pyrococcus horikoshii OT3 | | Descriptor: | hypothetical transcription regulator protein, PH1932 | | Authors: | Arai, R, Kishishita, S, Kukimoto-Niino, M, Wang, H, Sugawara, M, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-20 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical transcriptional regulator protein, PH1932 from Pyrococcus horikoshii OT3
To be Published
|
|
1X42
 
 | | Crystal structure of a haloacid dehalogenase family protein (PH0459) from Pyrococcus horikoshii OT3 | | Descriptor: | hypothetical protein PH0459 | | Authors: | Arai, R, Kukimoto-Niino, M, Sugahara, M, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-12 | | Release date: | 2005-11-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the probable haloacid dehalogenase PH0459 from Pyrococcus horikoshii OT3
Protein Sci., 15, 2006
|
|
1BVR
 
 | | M.TB. ENOYL-ACP REDUCTASE (INHA) IN COMPLEX WITH NAD+ AND C16-FATTY-ACYL-SUBSTRATE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE), TRANS-2-HEXADECENOYL-(N-ACETYL-CYSTEAMINE)-THIOESTER | | Authors: | Rozwarski, D.A, Vilcheze, C, Sugantino, M, Bittman, R, Jacobs, W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1998-09-17 | | Release date: | 1999-09-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis enoyl-ACP reductase, InhA, in complex with NAD+ and a C16 fatty acyl substrate.
J.Biol.Chem., 274, 1999
|
|
1ULR
 
 | | Crystal structure of tt0497 from Thermus thermophilus HB8 | | Descriptor: | putative acylphosphatase | | Authors: | Ago, H, Hamada, K, Sugahara, M, Kuroishi, C, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of tt0497 from Thermus thermophilus HB8
To be Published
|
|
1UJ4
 
 | | Crystal structure of Thermus thermophilus ribose-5-phosphate isomerase | | Descriptor: | CHLORIDE ION, ribose 5-phosphate isomerase | | Authors: | Hamada, K, Ago, H, Sugahara, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxyanion hole-stabilized stereospecific isomerization in ribose-5-phosphate isomerase (Rpi)
J.Biol.Chem., 278, 2003
|
|
1UJ6
 
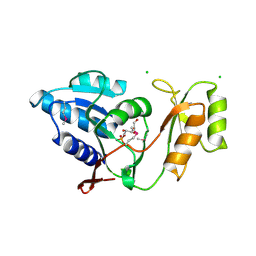 | | Crystal structure of Thermus thermophilus ribose-5-phosphate isomerase complexed with arabinose-5-phosphate | | Descriptor: | ARABINOSE-5-PHOSPHATE, CHLORIDE ION, ribose 5-phosphate isomerase | | Authors: | Hamada, K, Ago, H, Sugahara, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Oxyanion hole-stabilized stereospecific isomerization in ribose-5-phosphate isomerase (Rpi)
J.Biol.Chem., 278, 2003
|
|
1UJ5
 
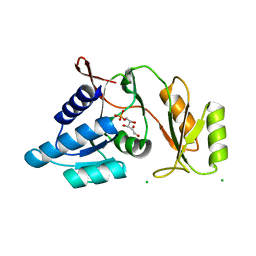 | | Crystal structure of Thermus thermophilus ribose-5-phosphate isomerase complexed with ribose-5-phosphate | | Descriptor: | CHLORIDE ION, RIBULOSE-5-PHOSPHATE, ribose 5-phosphate isomerase | | Authors: | Hamada, K, Ago, H, Sugahara, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxyanion hole-stabilized stereospecific isomerization in ribose-5-phosphate isomerase (Rpi)
J.Biol.Chem., 278, 2003
|
|
1V4R
 
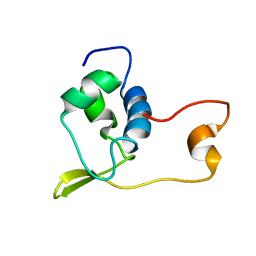 | | Solution structure of Streptmycal repressor TraR | | Descriptor: | Transcriptional Repressor | | Authors: | Tanaka, T, Komatsu, C, Kobayashi, K, Sugai, M, Kataoka, M, Kohno, T. | | Deposit date: | 2003-11-17 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Streptmycal repressor TraR
TO BE PUBLISHED
|
|
1ULS
 
 | | Crystal structure of tt0140 from Thermus thermophilus HB8 | | Descriptor: | putative 3-oxoacyl-acyl carrier protein reductase | | Authors: | Hisanaga, Y, Ago, H, Hamada, K, Sugahara, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of tt0140 from Thermus thermophilus HB8
To be Published
|
|
1WNF
 
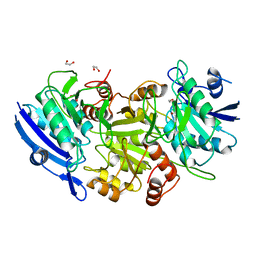 | | Crystal Structure of PH0066 from Pyrococcus horikoshii | | Descriptor: | 1,2-ETHANEDIOL, L-asparaginase | | Authors: | Ihsanawati, Sekine, S, Murayama, K, Sugawara, M, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-08-02 | | Release date: | 2005-02-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PH0066 from Pyrococcus horikoshii
To be Published
|
|
