2M33
 
 | | Solution NMR structure of full-length oxidized microsomal rabbit cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Subramanian, V, Ahuja, S, Popovych, N, Huang, R, Le Clair, S.V, Jahr, N, Soong, R, Xu, J, Yamamoto, K, Nanga, R.P, Im, S, Waskell, L, Ramamoorthy, A. | | Deposit date: | 2013-01-08 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of full-length mammalian cytochrome b5
To be Published
|
|
6GID
 
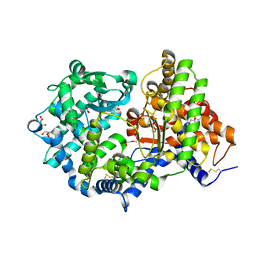 | | High resolution crystal structure of substrate-free human neprilysin | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moss, S, Subramanian, V, Acharya, K.R. | | Deposit date: | 2018-05-10 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution crystal structure of substrate-free human neprilysin.
J. Struct. Biol., 204, 2018
|
|
2ACF
 
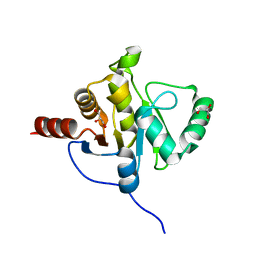 | | NMR STRUCTURE OF SARS-COV NON-STRUCTURAL PROTEIN NSP3A (SARS1) FROM SARS CORONAVIRUS | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab | | Authors: | Saikatendu, K.S, Joseph, J.S, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2005-07-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1''-phosphate dephosphorylation by a conserved domain of nsP3.
Structure, 13, 2005
|
|
2EPE
 
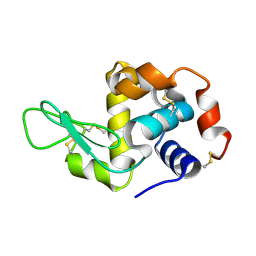 | | Crystal structure analysis of Hen egg white lysozyme grown by capillary method | | Descriptor: | Lysozyme C | | Authors: | Naresh, M.D, Subramanian, V, Jaimohan, S.M, Rajaram, A, Arumugam, V, Usha, R, Mandal, A.B. | | Deposit date: | 2007-03-29 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure analysis of Hen egg white lysozyme grown by capillary method
To be published
|
|
2BWK
 
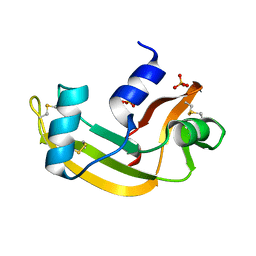 | | Murine angiogenin, sulphate complex | | Descriptor: | ANGIOGENIN, SULFATE ION | | Authors: | Holloway, D.E, Chavali, G.B, Hares, M.C, Subramanian, V, Acharya, K.R. | | Deposit date: | 2005-07-15 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Murine Angiogenin: Features of the Substrate- and Cell-Binding Regions and Prospects for Inhibitor-Binding Studies.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3B1U
 
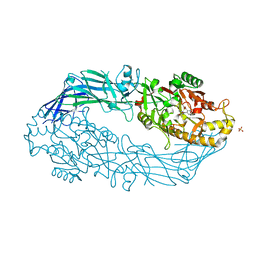 | | Crystal structure of human peptidylarginine deiminase 4 in complex with o-F-amidine | | Descriptor: | 2-{[(2S)-1-amino-5-{[(1Z)-2-fluoroethanimidoyl]amino}-1-oxopentan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Causey, C.P, Jones, J.E, Slack, J.L, Kamei, D, Jones Jr, L.E, Subramanian, V, Knuckley, B, Ebrahimi, P, Chumanevich, A.A, Luo, Y, Hashimoto, H, Shimizu, T, Sato, M, Hofseth, L.J, Thompson, P.R. | | Deposit date: | 2011-07-13 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Development of N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-fluoro-1-iminoethyl)-l-ornithine Amide (o-F-amidine) and N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-chloro-1-iminoethyl)-l-ornithine Amide (o-Cl-amidine) As Second Generation Protein Arginine Deiminase (PAD) Inhibitors
J.Med.Chem., 54, 2011
|
|
2BWL
 
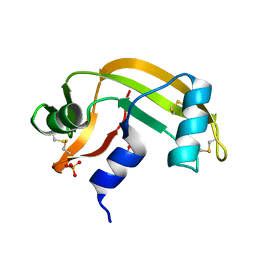 | | Murine angiogenin, phosphate complex | | Descriptor: | ANGIOGENIN, PHOSPHATE ION | | Authors: | Holloway, D.E, Chavali, G.B, Hares, M.C, Subramanian, V, Acharya, K.R. | | Deposit date: | 2005-07-15 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of Murine Angiogenin: Features of the Substrate- and Cell-Binding Regions and Prospects for Inhibitor-Binding Studies.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3B1T
 
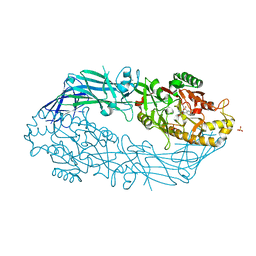 | | Crystal structure of human peptidylarginine deiminase 4 in complex with o-Cl-amidine | | Descriptor: | 2-{[(2S)-1-amino-5-{[(1Z)-2-chloroethanimidoyl]amino}-1-oxopentan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Causey, C.P, Jones, J.E, Slack, J.L, Kamei, D, Jones Jr, L.E, Subramanian, V, Knuckley, B, Ebrahimi, P, Chumanevich, A.A, Luo, Y, Hashimoto, H, Shimizu, T, Sato, M, Hofseth, L.J, Thompson, P.R. | | Deposit date: | 2011-07-13 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Development of N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-fluoro-1-iminoethyl)-l-ornithine Amide (o-F-amidine) and N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-chloro-1-iminoethyl)-l-ornithine Amide (o-Cl-amidine) As Second Generation Protein Arginine Deiminase (PAD) Inhibitors
J.Med.Chem., 54, 2011
|
|
4DKT
 
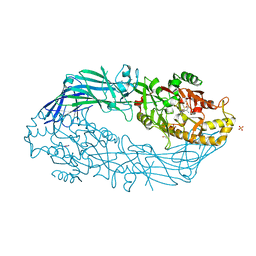 | | Crystal structure of human peptidylarginine deiminase 4 in complex with N-acetyl-L-threonyl-L-alpha-aspartyl-N5-[(1E)-2-fluoroethanimidoyl]-L-ornithinamide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Jones, J.E, Slack, J.L, Fang, P, Zhang, X, Subramanian, V, Causey, C.P, Coonrod, S.A, Guo, M, Thompson, P.R. | | Deposit date: | 2012-02-04 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Synthesis and Screening of a Haloacetamidine Containing Library To Identify PAD4 Selective Inhibitors.
Acs Chem.Biol., 7, 2012
|
|
2OG3
 
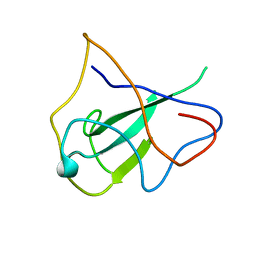 | | structure of the rna binding domain of n protein from SARS coronavirus in cubic crystal form | | Descriptor: | Nucleocapsid protein | | Authors: | Saikatendu, K, Joseph, J, Subramanian, V, Neuman, B, Buchmeier, M, Stevens, R.C, Kuhn, P. | | Deposit date: | 2007-01-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein.
J.Virol., 81, 2007
|
|
2OFZ
 
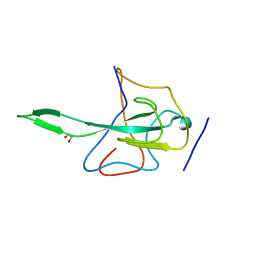 | | Ultrahigh Resolution Crystal Structure of RNA Binding Domain of SARS Nucleopcapsid (N Protein) at 1.1 Angstrom Resolution in Monoclinic Form. | | Descriptor: | 1,2-ETHANEDIOL, Nucleocapsid protein | | Authors: | Saikatendu, K, Joseph, J, Subramanian, V, Neuman, B, Buchmeier, M, Stevens, R.C, Kuhn, P. | | Deposit date: | 2007-01-04 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein.
J.Virol., 81, 2007
|
|
2OZK
 
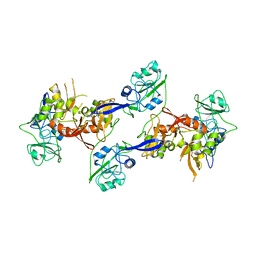 | | Structure of an N-Terminal Truncated Form of Nendou (NSP15) From SARS-CORONAVIRUS | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Saikatendu, K, Joseph, J, Subramanian, V, Neuman, B, Buchmeier, M, Stevens, R.C, Kuhn, P. | | Deposit date: | 2007-02-26 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch.
J.Virol., 81, 2007
|
|
6SH2
 
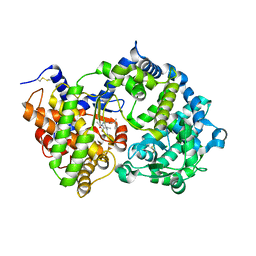 | | Crystal structure of human neprilysin E584D in complex with C-type natriuretic peptide. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type natriuretic peptide fragment (CNP), CHLORIDE ION, ... | | Authors: | Moss, S, Subramanian, V, Acharya, K.R. | | Deposit date: | 2019-08-05 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of peptide-bound neprilysin reveals key binding interactions.
Febs Lett., 594, 2020
|
|
6SH1
 
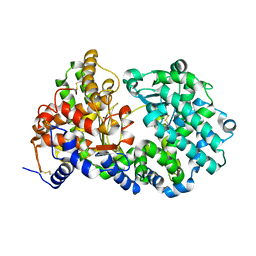 | | Crystal structure of substrate-free human neprilysin E584D. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, ... | | Authors: | Moss, S, Subramanian, V, Acharya, K.R. | | Deposit date: | 2019-08-05 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of peptide-bound neprilysin reveals key binding interactions.
Febs Lett., 594, 2020
|
|
2VQ9
 
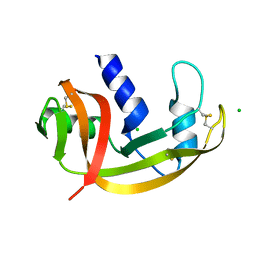 | | RNASE ZF-3E | | Descriptor: | CHLORIDE ION, RNASE 1 | | Authors: | Kazakou, K, Holloway, D.E, Prior, S.H, Subramanian, V, Acharya, K.R. | | Deposit date: | 2008-03-12 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ribonuclease A Homologues of the Zebrafish: Polymorphism, Crystal Structures of Two Representatives and Their Evolutionary Implications
J.Mol.Biol., 380, 2008
|
|
2VQ8
 
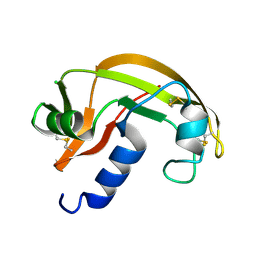 | | RNASE ZF-1A | | Descriptor: | CHLORIDE ION, RNASE ZF-1A | | Authors: | Kazakou, K, Holloway, D.E, Prior, S.H, Subramanian, V, Acharya, K.R. | | Deposit date: | 2008-03-12 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Ribonuclease A Homologues of the Zebrafish: Polymorphism, Crystal Structures of Two Representatives and Their Evolutionary Implications.
J.Mol.Biol., 380, 2008
|
|
2IDY
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Horst, R, Herrmann, T, Joseph, J, Saikatendu, K, Subramanian, V, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2GRI
 
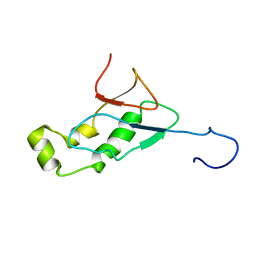 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Herrmann, T, Saikatendu, K.S, Joseph, J, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-04-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
2J4T
 
 | | Biological and Structural Features of Murine Angiogenin-4, an Angiogenic Protein | | Descriptor: | ANGIOGENIN-4 | | Authors: | Crabtree, B, Holloway, D.E, Baker, M.D, Acharya, K.R, Subramanian, V. | | Deposit date: | 2006-09-05 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biological and Structural Features of Murine Angiogenin-4, an Angiogenic Protein
Biochemistry, 46, 2007
|
|
2KJ7
 
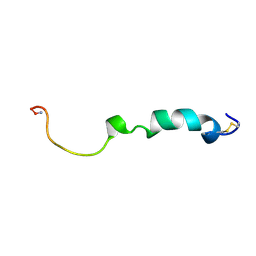 | | Three-Dimensional NMR Structure of Rat Islet Amyloid Polypeptide in DPC micelles | | Descriptor: | Islet amyloid polypeptide | | Authors: | Nanga, R, Brender, J.R, Xu, J, Hartman, K, Subramanian, V, Ramamoorthy, A. | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and orientation of rat islet amyloid polypeptide protein in a membrane environment by solution NMR spectroscopy.
J.Am.Chem.Soc., 131, 2009
|
|
2FYG
 
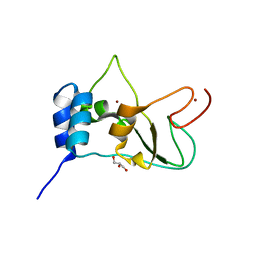 | | Crystal structure of NSP10 from Sars coronavirus | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab, ZINC ION | | Authors: | Joseph, J.S, Saikatendu, K.S, Subramanian, V, Neuman, B.W, Brooun, A, Griffith, M, Moy, K, Yadav, M.K, Velasquez, J, Buchmeier, M.J, Stevens, R.C, Kuhn, P. | | Deposit date: | 2006-02-07 | | Release date: | 2006-08-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs.
J.Virol., 80, 2006
|
|
4AHL
 
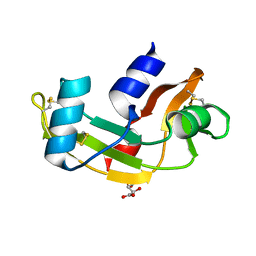 | | P112L - Angiogenin mutants and amyotrophic lateral sclerosis - a biochemical and biological analysis | | Descriptor: | ANGIOGENIN, L(+)-TARTARIC ACID | | Authors: | Thiyagarajan, N, Ferguson, R, Saha, S, Pham, T, Subramanian, V, Acharya, K.R. | | Deposit date: | 2012-02-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Structural and Molecular Insights Into the Mechanism of Action of Human Angiogenin-Als Variants in Neurons.
Nat.Commun., 3, 2012
|
|
4AHG
 
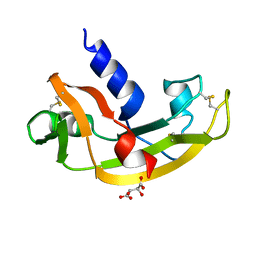 | | S28N - Angiogenin mutants and amyotrophic lateral sclerosis - a biochemical and biological analysis | | Descriptor: | ANGIOGENIN, D(-)-TARTARIC ACID | | Authors: | Thiyagarajan, N, Ferguson, R, Saha, S, Pham, T, Subramanian, V, Acharya, K.R. | | Deposit date: | 2012-02-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.448 Å) | | Cite: | Structural and Molecular Insights Into the Mechanism of Action of Human Angiogenin-Als Variants in Neurons.
Nat.Commun., 3, 2012
|
|
4AHK
 
 | | K54E - Angiogenin mutants and amyotrophic lateral sclerosis - a biochemical and biological analysis | | Descriptor: | ANGIOGENIN, D(-)-TARTARIC ACID | | Authors: | Thiyagarajan, N, Ferguson, R, Saha, S, Pham, T, Subramanian, V, Acharya, K.R. | | Deposit date: | 2012-02-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Molecular Insights Into the Mechanism of Action of Human Angiogenin-Als Variants in Neurons.
Nat.Commun., 3, 2012
|
|
4AOH
 
 | | Structural snapshots and functional analysis of human angiogenin variants associated with Amyotrophic Lateral Sclerosis (ALS) | | Descriptor: | ANGIOGENIN, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID | | Authors: | Thiyagarajan, N, Ferguson, R, Subramanian, V, Acharya, K.R. | | Deposit date: | 2012-03-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.041 Å) | | Cite: | Structural and Molecular Insights Into the Mechanism of Action of Human Angiogenin-Als Variants in Neurons
Nat.Commun., 3, 2012
|
|
