3NRY
 
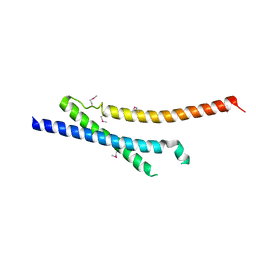 | | Insights into anti-parallel microtubule crosslinking by PRC1, a conserved microtubule binding protein | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Kapoor, T.M, Subramanian, R, Wilson-Kubalek, E.M, Arthur, C.P, Bick, M.J, Campbell, E.A, Darst, S.A, Milligan, R.A. | | Deposit date: | 2010-07-01 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into Antiparallel Microtubule Crosslinking by PRC1, a Conserved Nonmotor Microtubule Binding Protein.
Cell(Cambridge,Mass.), 142, 2010
|
|
6LVB
 
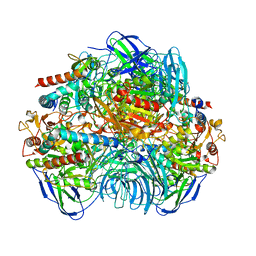 | | Structure of Dimethylformamidase, tetramer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LAY
 
 | |
5ZA4
 
 | |
6LVC
 
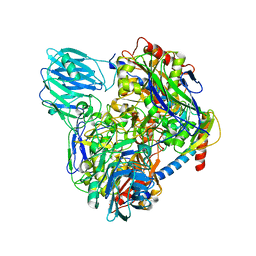 | | Structure of Dimethylformamidase, dimer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6MLR
 
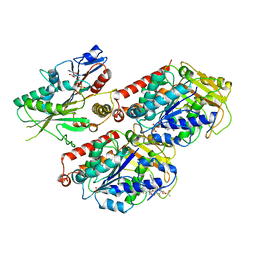 | | Cryo-EM structure of microtubule-bound Kif7 in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF7, ... | | Authors: | Mani, N, Jiang, S, Wilson-Kubalek, E.M, Ku, P, Milligan, R.A, Subramanian, R. | | Deposit date: | 2018-09-27 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Interplay between the Kinesin and Tubulin Mechanochemical Cycles Underlies Microtubule Tip Tracking by the Non-motile Ciliary Kinesin Kif7.
Dev.Cell, 49, 2019
|
|
6MLQ
 
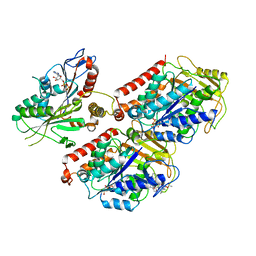 | | Cryo-EM structure of microtubule-bound Kif7 in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mani, N, Jiang, S, Wilson-Kubalek, E.M, Ku, P, Milligan, R.A, Subramanian, R. | | Deposit date: | 2018-09-27 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Interplay between the Kinesin and Tubulin Mechanochemical Cycles Underlies Microtubule Tip Tracking by the Non-motile Ciliary Kinesin Kif7.
Dev.Cell, 49, 2019
|
|
6LVD
 
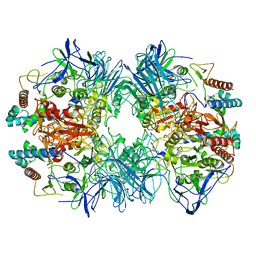 | | Structure of Dimethylformamidase, tetramer, Y440A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LVE
 
 | | Structure of Dimethylformamidase, tetramer, E521A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6B8F
 
 | | Contracted Human Heavy-Chain Ferritin Crystal-Hydrogel Hybrid | | Descriptor: | CALCIUM ION, FE (III) ION, Ferritin heavy chain | | Authors: | Zhang, L, Bailey, J.B, Subramanian, R, Tezcan, F.A. | | Deposit date: | 2017-10-07 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Hyperexpandable, self-healing macromolecular crystals with integrated polymer networks.
Nature, 557, 2018
|
|
6B8G
 
 | | Twice-Contracted Human Heavy-Chain Ferritin Crystal-Hydrogel Hybrid | | Descriptor: | CALCIUM ION, FE (III) ION, Ferritin heavy chain | | Authors: | Zhang, L, Bailey, J.B, Subramanian, R, Tezcan, F.A. | | Deposit date: | 2017-10-07 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Hyperexpandable, self-healing macromolecular crystals with integrated polymer networks.
Nature, 557, 2018
|
|
5F6Z
 
 | | Sandercyanin Fluorescent Protein purified from Sander vitreus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, Sandercyanin Fluorescent Protein | | Authors: | Ghosh, S, Yu, C.L, Ferraro, D, Sudha, S, Pal, S, Schaefer, W, Gibson, D.T, Subramanian, R. | | Deposit date: | 2015-12-07 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Blue protein with red fluorescence
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8G4V
 
 | | Horse liver alcohol dehydrogense His-51-Gln form complexed with NAD+ and 2,3,4,5,6-pentafluorobenzyl alcohol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V, Subramanian, R. | | Deposit date: | 2023-02-10 | | Release date: | 2023-02-22 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Histidine-51 facilitates deprotonation of the zinc-bound ligand during catalysis by horse liver alcohol dehydrogenase
To Be Published
|
|
5ZJM
 
 | | Crystal structure of N-acetylneuraminate lyase from Fusobacterium nucleatum | | Descriptor: | 1,2-ETHANEDIOL, N-acetylneuraminate lyase | | Authors: | Kumar, J.P, Rao, H, Nayak, V, Subramanian, R. | | Deposit date: | 2018-03-21 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.323 Å) | | Cite: | Crystal structures and kinetics of N-acetylneuraminate lyase from Fusobacterium nucleatum
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
