3C0K
 
 | |
1FBB
 
 | |
4GA0
 
 | |
8I4J
 
 | |
1FBK
 
 | |
4GA2
 
 | |
8I4K
 
 | | Structure of Azami Red1.0, a red fluorescent protein engineered from Azami Green | | Descriptor: | Azami Red1.0, CALCIUM ION | | Authors: | Otsubo, S, Takekawa, N, Imamura, H, Imada, K. | | Deposit date: | 2023-01-19 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Red fluorescent proteins engineered from green fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G1R
 
 | |
4BK0
 
 | | Crystal structure of the KIX domain of human RECQL5 (domain-swapped dimer) | | Descriptor: | ATP-DEPENDENT DNA HELICASE Q5, DI(HYDROXYETHYL)ETHER | | Authors: | Kassube, S.A, Jinek, M, Fang, J, Tsutakawa, S, Nogales, E. | | Deposit date: | 2013-04-21 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Mimicry in Transcription Regulation of Human RNA Polymerase II by the DNA Helicase Recql5
Nat.Struct.Mol.Biol., 20, 2013
|
|
5VX6
 
 | | Structure of Bacillus subtilis Inhibitor of motility (MotI/DgrA) | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Uncharacterized protein YpfA | | Authors: | Subramanian, S, Dann III, C. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | MotI (DgrA) acts as a molecular clutch on the flagellar stator protein MotA inBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6TBL
 
 | |
6TBN
 
 | | Crystal structure of CIAO1-CIAO2B CIA core complex | | Descriptor: | MIP18 family protein galla-2, Probable cytosolic iron-sulfur protein assembly protein Ciao1, SODIUM ION | | Authors: | Kassube, S.A, Thoma, N.H. | | Deposit date: | 2019-11-01 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into Fe-S protein biogenesis by the CIA targeting complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6TC0
 
 | | Crystal structure of MMS19-CIAO1-CIAO2B CIA targeting complex | | Descriptor: | MIP18 family protein galla-2, MMS19 nucleotide excision repair protein homolog, Probable cytosolic iron-sulfur protein assembly protein Ciao1 | | Authors: | Kassube, S.A, Thoma, N.H. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insights into Fe-S protein biogenesis by the CIA targeting complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8ELD
 
 | |
8EM6
 
 | |
8ES4
 
 | |
6CVM
 
 | | Atomic resolution cryo-EM structure of beta-galactosidase | | Descriptor: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, MAGNESIUM ION, ... | | Authors: | Subramaniam, S, Bartesaghi, A, Banerjee, S, Zhu, X, Milne, J.L.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Atomic Resolution Cryo-EM Structure of beta-Galactosidase.
Structure, 26, 2018
|
|
6WQH
 
 | | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ig2 substrate, Lon protease, ... | | Authors: | Zhang, K, Li, S, Hsiehb, K, Sub, S, Pintilie, G, Chiu, W, Chang, C. | | Deposit date: | 2020-04-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
J.Biol.Chem., 297, 2021
|
|
4UQQ
 
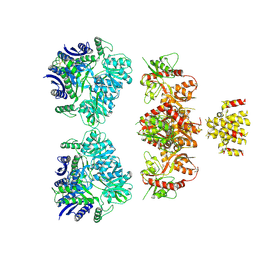 | | Electron density map of GluK2 desensitized state in complex with 2S,4R-4-methylglutamate | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, KAINATE 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQJ
 
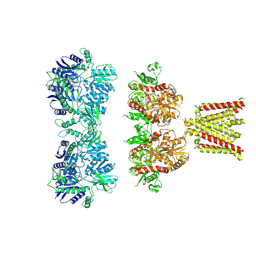 | | Cryo-EM density map of GluA2em in complex with ZK200775 | | Descriptor: | GLUTAMATE RECEPTOR 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
1R69
 
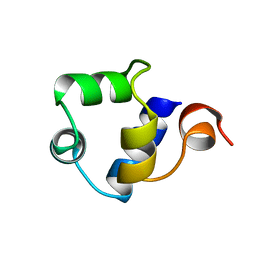 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | Deposit date: | 1988-12-08 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
4UQK
 
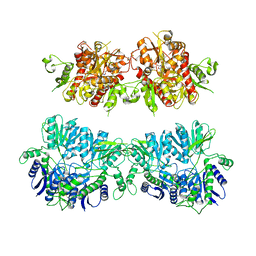 | | Electron density map of GluA2em in complex with quisqualate and LY451646 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2 | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
6EBM
 
 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, transmembrane domain of subunit alpha | | Descriptor: | Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6UAN
 
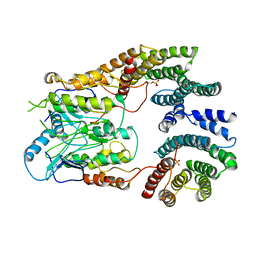 | | B-Raf:14-3-3 complex | | Descriptor: | 14-3-3 zeta, Serine/threonine-protein kinase B-raf | | Authors: | Kondo, Y, Ognjenovic, J, Banerjee, S, Karandur, D, Merk, A, Kulhanek, K, Wong, K, Roose, J.P, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2019-09-11 | | Release date: | 2019-09-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases.
Science, 366, 2019
|
|
3JCG
 
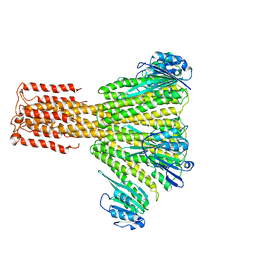 | |
