3BR8
 
 | |
2HCU
 
 | | Crystal Structure Of Smu.1381 (or LeuD) from Streptococcus Mutans | | Descriptor: | 3-isopropylmalate dehydratase small subunit, SULFATE ION | | Authors: | Gao, Z.Q, Hou, H.F, Li, L.F, Liang, Y.H, Su, X.D, Dong, Y.H. | | Deposit date: | 2006-06-19 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure Of Smu.1381 (or LeuD) from Streptococcus Mutans
To be Published
|
|
3BYD
 
 | | Crystal structure of beta-lactamase OXY-1-1 from Klebsiella oxytoca | | Descriptor: | ACETATE ION, Beta-lactamase OXY-1, SULFATE ION | | Authors: | Liang, Y.-H, Wu, S.W, Su, X.-D. | | Deposit date: | 2008-01-15 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural insights into the broadened substrate profile of the extended-spectrum beta-lactamase OXY-1-1 from Klebsiella oxytoca
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3B3D
 
 | | B.subtilis YtbE | | Descriptor: | CALCIUM ION, Putative morphine dehydrogenase | | Authors: | Zhou, Y.F, Li, L.F, Liang, Y.H, Su, X.-D. | | Deposit date: | 2007-10-20 | | Release date: | 2008-10-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical analyses of YvgN and YtbE from Bacillus subtilis
Protein Sci., 18, 2009
|
|
7BUM
 
 | | mcGAS bound with pGpA | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cyclic GMP-AMP synthase, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Wang, B, Su, X.D. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.047 Å) | | Cite: | Mn2+Directly Activates cGAS and Structural Analysis Suggests Mn2+Induces a Noncanonical Catalytic Synthesis of 2'3'-cGAMP.
Cell Rep, 32, 2020
|
|
7BUQ
 
 | | mcGAS bound with 23-cGAMP | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION, cGAMP | | Authors: | Wang, B, Su, X.D. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Mn2+Directly Activates cGAS and Structural Analysis Suggests Mn2+Induces a Noncanonical Catalytic Synthesis of 2'3'-cGAMP.
Cell Rep, 32, 2020
|
|
7QA9
 
 | | 10bp DNA/DNA duplex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*TP*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | Authors: | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7BUJ
 
 | | mcGAS bound with pppGpG | | Descriptor: | Cyclic GMP-AMP synthase, GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, B, Su, X.D. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Mn2+Directly Activates cGAS and Structural Analysis Suggests Mn2+Induces a Noncanonical Catalytic Synthesis of 2'3'-cGAMP.
Cell Rep, 32, 2020
|
|
3EXT
 
 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | MAGNESIUM ION, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Liu, X, Li, G.L, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
7EK0
 
 | | Complex Structure of antibody BD-503 and RBD-N501Y of COVID-19 | | Descriptor: | Heavy Chain of BD-503, Light Chain of BD-503, Spike protein S1 | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7EJZ
 
 | | Complex Structure of antibody BD-503 and RBD-S477N of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7EJY
 
 | | Complex Structure of antibody BD-503 and RBD of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
3EXR
 
 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3EXS
 
 | | Crystal structure of KGPDC from Streptococcus mutans in complex with D-R5P | | Descriptor: | RIBULOSE-5-PHOSPHATE, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Wang, K.T, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3JCU
 
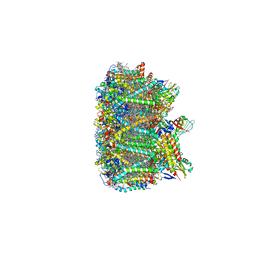 | | Cryo-EM structure of spinach PSII-LHCII supercomplex at 3.2 Angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Wei, X.P, Zhang, X.Z, Su, X.D, Cao, P, Liu, X.Y, Li, M, Chang, W.R, Liu, Z.F. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of spinach photosystem II-LHCII supercomplex at 3.2 A resolution
Nature, 534, 2016
|
|
7F6Z
 
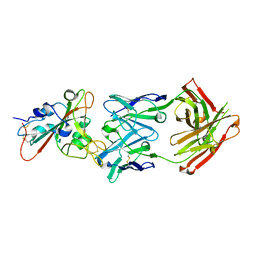 | | Complex Structure of antibody BD-503 and RBD-501Y.V2 of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-06-26 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7F6Y
 
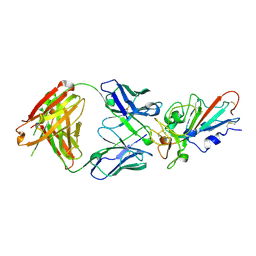 | | Complex Structure of antibody BD-503 and RBD-E484K of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-06-26 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
3GAD
 
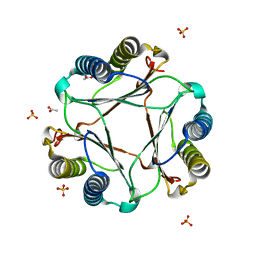 | | Structure of apomif | | Descriptor: | ACETIC ACID, Macrophage migration inhibitory factor-like protein, SULFATE ION | | Authors: | Zhou, Y.-F, Su, X.-D, Shao, D, Wang, H. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional comparison of MIF ortholog from Plasmodium yoelii with MIF from its rodent host
Mol.Immunol., 47, 2010
|
|
3GAC
 
 | | Structure of mif with HPP | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ACETIC ACID, Macrophage migration inhibitory factor-like protein, ... | | Authors: | Zhou, Y.-F, Su, X.-D, Shao, D, Wang, H. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional comparison of MIF ortholog from Plasmodium yoelii with MIF from its rodent host
Mol.Immunol., 47, 2010
|
|
3H6X
 
 | |
3E4O
 
 | | Crystal structure of succinate bound state DctB | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, MAGNESIUM ION, SUCCINIC ACID | | Authors: | Zhou, Y.F, Nan, J, Nan, B.Y, Liang, Y.H, Panjikar, S, Su, X.D. | | Deposit date: | 2008-08-12 | | Release date: | 2008-10-21 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain.
J.Mol.Biol., 383, 2008
|
|
3RNY
 
 | | Crystal structure of human RSK1 C-terminal kinase domain | | Descriptor: | Ribosomal protein S6 kinase alpha-1, SODIUM ION | | Authors: | Li, D, Fu, T.-M, Nan, J, Su, X.-D. | | Deposit date: | 2011-04-24 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the autoinhibition of the C-terminal kinase domain of human RSK1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4O9I
 
 | |
3L7P
 
 | |
3L7T
 
 | |
