8HCT
 
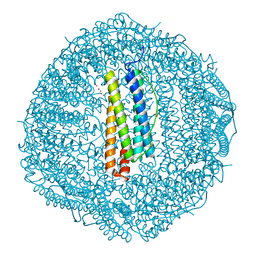 | | Crystal structure of Cu2+ binding to Dendrorhynchus zhejiangensis ferritin | | Descriptor: | COPPER (II) ION, FE (III) ION, Ferritin, ... | | Authors: | Ming, T.H, Su, X.R, Huo, C.H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural and Biochemical Characterization of Silver/Copper Binding by Dendrorhynchus zhejiangensis Ferritin.
Polymers (Basel), 15, 2023
|
|
1LM0
 
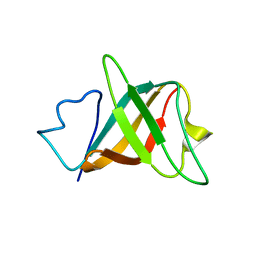 | | Solution structure and characterization of the heme chaperone CcmE | | Descriptor: | cytochrome c maturation protein E | | Authors: | Arnesano, F, Banci, L, Barker, P.D, Bertini, I, Rosato, A, Su, X.C, Viezzoli, M.S. | | Deposit date: | 2002-04-30 | | Release date: | 2002-12-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and characterization of the heme chaperone CcmE
Biochemistry, 41, 2002
|
|
1P6T
 
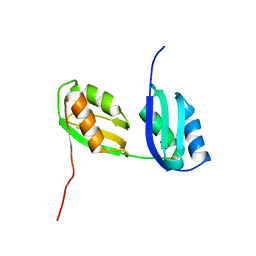 | | Structure characterization of the water soluble region of P-type ATPase CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, L, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-04-30 | | Release date: | 2003-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the function of the N-terminal domain of the ATPase CopA from Bacillus subtilis.
J.Biol.Chem., 278, 2003
|
|
1OQ3
 
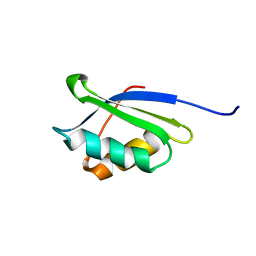 | | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, L, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-07 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis
J.Mol.Biol., 331, 2003
|
|
1OPZ
 
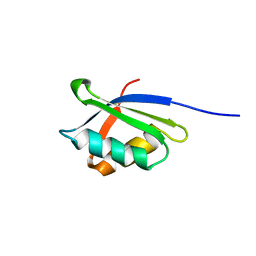 | | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonneli, L, Su, X.C. | | Deposit date: | 2003-03-06 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis.
J.Mol.Biol., 331, 2003
|
|
1OQ6
 
 | | solution structure of Copper-S46V CopA from Bacillus subtilis | | Descriptor: | COPPER (II) ION, Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, l, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-07 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis
J.Mol.Biol., 331, 2003
|
|
7QA9
 
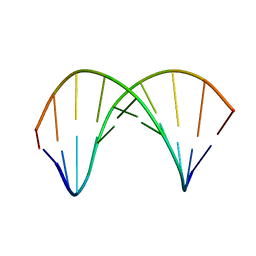 | | 10bp DNA/DNA duplex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*TP*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | Authors: | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1HXJ
 
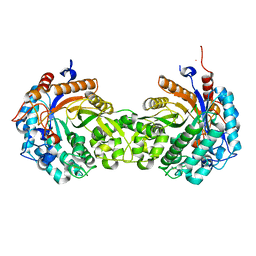 | |
7VHR
 
 | | Apostichopus japonicus ferritin | | Descriptor: | Ferritin, MAGNESIUM ION | | Authors: | Wu, Y, Su, X.R, Ming, T.H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Crystallographic characterization of a marine invertebrate ferritin from the sea cucumber Apostichopus japonicus.
Febs Open Bio, 12, 2022
|
|
1KTV
 
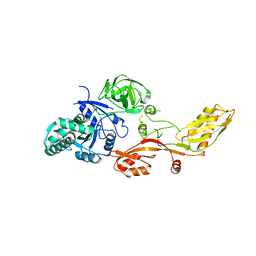 | |
1SB6
 
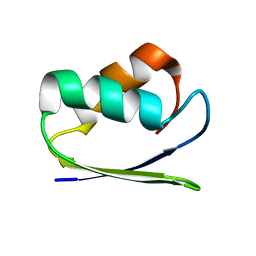 | | Solution structure of a cyanobacterial copper metallochaperone, ScAtx1 | | Descriptor: | copper chaperone ScAtx1 | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Su, X.C, Borrelly, G.P, Robinson, N.J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-02-10 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a Cyanobacterial Metallochaperone: INSIGHT INTO AN ATYPICAL COPPER-BINDING MOTIF.
J.Biol.Chem., 279, 2004
|
|
2K7R
 
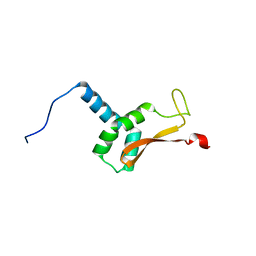 | | N-terminal domain of the Bacillus subtilis helicase-loading protein DnaI | | Descriptor: | Primosomal protein dnaI, ZINC ION | | Authors: | Loscha, K.V, Jaudzems, K, Ioannou, C, Su, X.C, Hill, F.R, Otting, G, Dixon, N.E, Liepinsh, E. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader
Nucleic Acids Res., 37, 2009
|
|
2AJ1
 
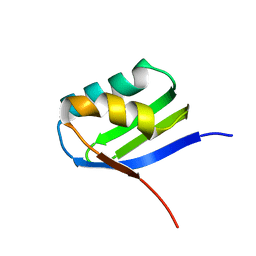 | | Solution structure of apoCadA | | Descriptor: | Probable cadmium-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Su, X.-C, Miras, R, Bal, N, Mintz, E, Catty, P, Shokes, J.E, Scott, R.A. | | Deposit date: | 2005-08-01 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for metal binding specificity: the N-terminal cadmium binding domain of the P1-type ATPase CadA
J.Mol.Biol., 356, 2006
|
|
2AJ0
 
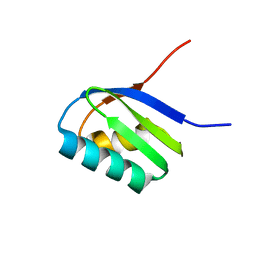 | | Solution structure of apoCadA | | Descriptor: | Probable cadmium-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Su, X.-C, Miras, R, Bal, N, Mintz, E, Catty, P, Shokes, J.E, Scott, R.A. | | Deposit date: | 2005-08-01 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for metal binding specificity: the N-terminal cadmium binding domain of the P1-type ATPase CadA
J.Mol.Biol., 356, 2006
|
|
2AYA
 
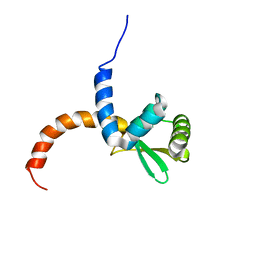 | |
1XIK
 
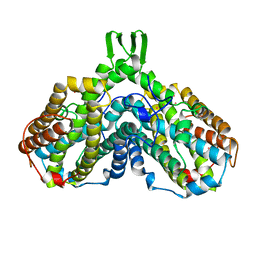 | | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 BETA CHAIN | | Descriptor: | FE (II) ION, MERCURY (II) ION, PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE | | Authors: | Logan, D.T, Su, X.-D, Aberg, A, Regnstrom, K, Hajdu, J, Eklund, H, Nordlund, P. | | Deposit date: | 1996-08-06 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of reduced protein R2 of ribonucleotide reductase: the structural basis for oxygen activation at a dinuclear iron site.
Structure, 4, 1996
|
|
1PFR
 
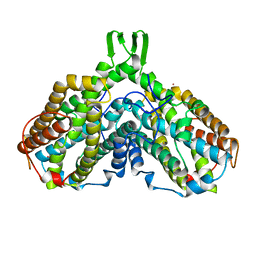 | | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 BETA CHAIN | | Descriptor: | FE (III) ION, MERCURY (II) ION, PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE | | Authors: | Logan, D.T, Su, X.D, Aberg, A, Regnstrom, K, Hajdu, J, Eklund, H, Nordlund, P. | | Deposit date: | 1996-12-03 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of reduced protein R2 of ribonucleotide reductase: the structural basis for oxygen activation at a dinuclear iron site.
Structure, 4, 1996
|
|
1RN7
 
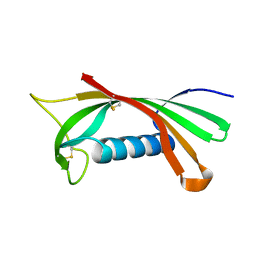 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-11-30 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
4E2A
 
 | |
3CP7
 
 | |
1ROA
 
 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-12-01 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
5SY7
 
 | | Crystal Structure of the Heterodimeric NPAS3-ARNT Complex with HRE DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*CP*AP*CP*GP*AP*CP*CP*CP*GP*CP*AP*CP*GP*TP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*GP*CP*GP*TP*AP*CP*GP*TP*GP*CP*GP*GP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Wu, D, Su, X, Potluri, N, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors.
Elife, 5, 2016
|
|
5SY5
 
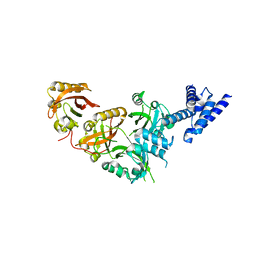 | | Crystal Structure of the Heterodimeric NPAS1-ARNT Complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Neuronal PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Potluri, N, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors.
Elife, 5, 2016
|
|
3D3F
 
 | |
2D4G
 
 | |
