5LQ3
 
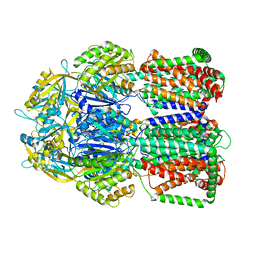 | |
8EL9
 
 | | Cryo-EM structure of human catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Su, C.C. | | Deposit date: | 2022-09-23 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Cryo-EM structure of human catalase
To Be Published
|
|
8T3U
 
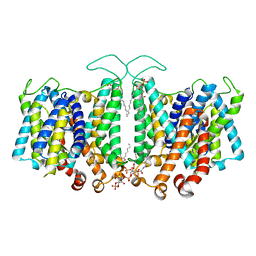 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaCl Buffer: Form2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
8T45
 
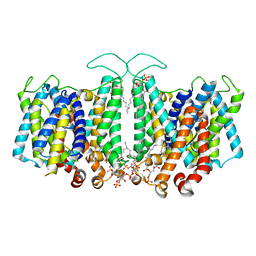 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaHCO3 Buffer: Form2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
8T47
 
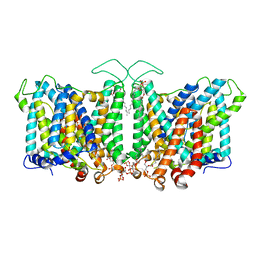 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaHCO3 Buffer: Form3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CARBONATE ION, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
8T44
 
 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaHCO3 Buffer: Form1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CARBONATE ION, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
8T3R
 
 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaCl Buffer: Form1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CHLORIDE ION, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
7N6B
 
 | | Structure of MmpL3 reconstituted into lipid nanodisc in the TMM bound state | | Descriptor: | 6-O-[(2S)-2-{(1S)-18-[(1R,2R)-2-hexylcyclopropyl]-1-hydroxyoctadecyl}tricosanoyl]-alpha-D-glucopyranosyl alpha-D-glucopyranoside, MmpL3 transporter | | Authors: | Su, C.C, Yu, E. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structures of the mycobacterial membrane protein MmpL3 reveal its mechanism of lipid transport.
Plos Biol., 19, 2021
|
|
8EKY
 
 | | Cryo-EM structure of the human PRDX4-ErP46 complex | | Descriptor: | Peroxiredoxin-4, Thioredoxin domain-containing protein 5 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EKW
 
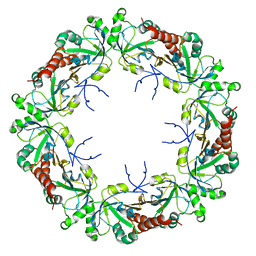 | | Cryo-EM structure of human PRDX4 | | Descriptor: | Peroxiredoxin-4 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EM2
 
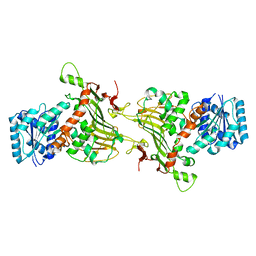 | |
9BFM
 
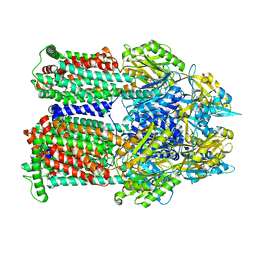 | | Cryo-EM co-structure of AcrB with the EPM35 efflux pump inhibitor | | Descriptor: | (2S)-1-(3,4-dichlorophenoxy)-3-(4-{[4-(trifluoromethyl)pyrimidin-2-yl]amino}piperidin-1-yl)propan-2-ol, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
9BFH
 
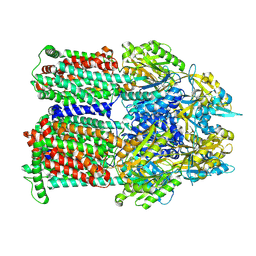 | | Cryo-EM co-structure of AcrB with the CU032 efflux pump inhibitor | | Descriptor: | (2S)-1-[(3R)-3-aminopyrrolidin-1-yl]-3-(3,4-dichlorophenoxy)propan-2-ol, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-17 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
9BFT
 
 | | Cryo-EM co-structure of AcrB with CU244 | | Descriptor: | (2S)-1-{[(1R,5R)-3-azabicyclo[3.1.0]hexan-6-yl]amino}-3-(3,5-dichlorophenoxy)propan-2-ol, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
9BFN
 
 | | Cryo-EM co-structure of AcrB with the CU232 efflux pump inhibitor | | Descriptor: | (2R)-1-(4-aminopiperidin-1-yl)-3-[3-(trifluoromethyl)phenoxy]propan-2-ol, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Multidrug efflux pump subunit AcrB | | Authors: | Su, C.C. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Bacterial efflux pump modulators prevent bacterial growth in macrophages and under broth conditions that mimic the host environment.
mBio, 14, 2023
|
|
6VQQ
 
 | |
6VQR
 
 | | CryoEM Structure of the PfFNT-inhibitor complex | | Descriptor: | (2R)-2-hydroxy-7-methoxy-2-(pentafluoroethyl)-2,3-dihydro-4H-1-benzopyran-4-one, Formate-nitrite transporter | | Authors: | Su, C.C, Lyu, M. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural basis of transport and inhibition of the Plasmodium falciparum transporter PfFNT.
Embo Rep., 22, 2021
|
|
6E5D
 
 | |
6E5F
 
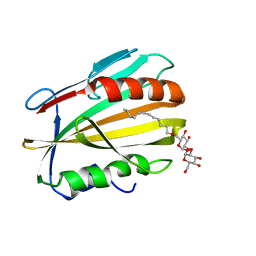 | |
6MNA
 
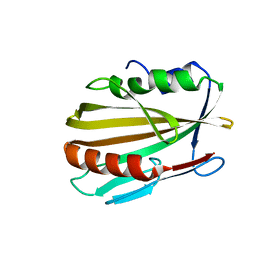 | |
3BCG
 
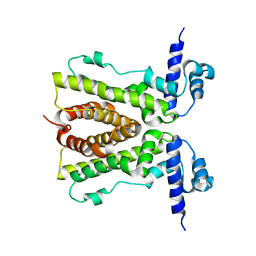 | | Conformational changes of the AcrR regulator reveal a mechanism of induction | | Descriptor: | HTH-type transcriptional regulator acrR | | Authors: | Gu, R, Li, M, Su, C.C, Long, F, Yang, F, McDermott, G, Yu, E.Y. | | Deposit date: | 2007-11-12 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conformational change of the AcrR regulator reveals a possible mechanism of induction.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3QPS
 
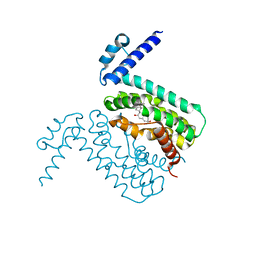 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
