5D1W
 
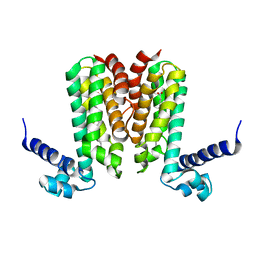 | | Crystal structure of Mycobacterium tuberculosis Rv3249c transcriptional regulator. | | Descriptor: | PALMITIC ACID, Rv3249c transcriptional regulator | | Authors: | Chou, T.-H, Delmar, J, Su, C.-C, Yu, E. | | Deposit date: | 2015-08-04 | | Release date: | 2015-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural Basis for the Regulation of the MmpL Transporters of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
7JZ3
 
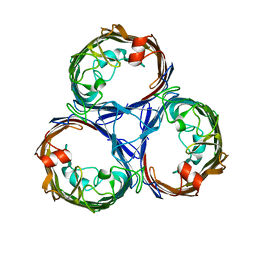 | | Osmoporin OmpC from E.coli K12 | | Descriptor: | Outer membrane protein C | | Authors: | Lyu, M, Su, C, Morgan, C.E, Bolla, J.R, Robinson, C.V, Yu, E.W. | | Deposit date: | 2020-09-01 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
2QOP
 
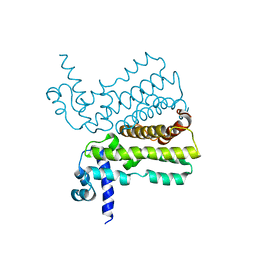 | | Crystal structure of the transcriptional regulator AcrR from Escherichia coli | | Descriptor: | HTH-type transcriptional regulator acrR | | Authors: | Li, M, Gu, R, Su, C.-C, McDermott, G, Yu, E.W. | | Deposit date: | 2007-07-20 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the transcriptional regulator AcrR from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
6WU6
 
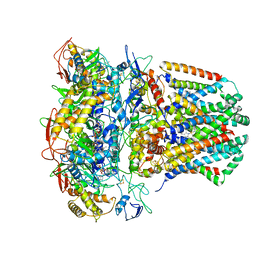 | | succinate-coenzyme Q reductase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Lyu, M, Su, C.-C, Morgan, C.E, Yu, E.W. | | Deposit date: | 2020-05-04 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
6WTZ
 
 | | Cryo-EM structure of E. Coli OmpF | | Descriptor: | Outer membrane porin F | | Authors: | Morgan, C.E, Su, C.-C, Lyu, M, Yu, E.W. | | Deposit date: | 2020-05-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
2QCO
 
 | | Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni | | Descriptor: | CmeR, GLYCEROL | | Authors: | Gu, R, Su, C, Shi, F, Li, M, McDermott, G, Zhang, Q, Yu, E.W. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni.
J.Mol.Biol., 372, 2007
|
|
3QQA
 
 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.-C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
6MNA
 
 | |
5D9R
 
 | | Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6 | | Descriptor: | Protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic | | Authors: | Radhakrishnan, A, Kumar, N, Su, C.-C, Chou, T.-H, Yu, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6.
Protein Sci., 25, 2016
|
|
7JZ2
 
 | | Succinate: quinone oxidoreductase SQR from E.coli K12 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lyu, M, Su, C.-C, Morgan, C.E, Bolla, J.R, Robinson, C.V, Yu, E.W. | | Deposit date: | 2020-09-01 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
3BCG
 
 | | Conformational changes of the AcrR regulator reveal a mechanism of induction | | Descriptor: | HTH-type transcriptional regulator acrR | | Authors: | Gu, R, Li, M, Su, C.C, Long, F, Yang, F, McDermott, G, Yu, E.Y. | | Deposit date: | 2007-11-12 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conformational change of the AcrR regulator reveals a possible mechanism of induction.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3QPS
 
 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
7EKE
 
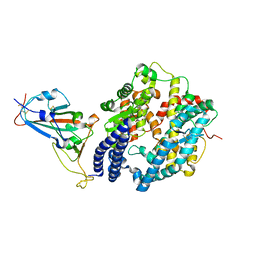 | | Structure of SARS-CoV-2 spike receptor-binding domain F486L mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKC
 
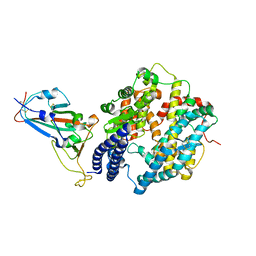 | | Structure of SARS-CoV-2 Gamma variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKG
 
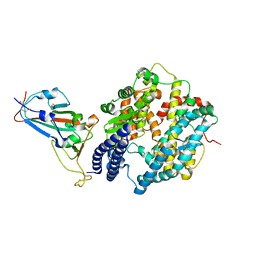 | | Structure of SARS-CoV-2 Beta variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKH
 
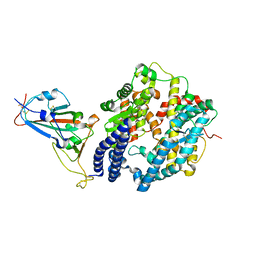 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKF
 
 | | Structure of SARS-CoV-2 Alpha variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
3R24
 
 | | Crystal structure of nsp10/nsp16 complex of SARS coronavirus | | Descriptor: | 2'-O-methyl transferase, Non-structural protein 10 and Non-structural protein 11, S-ADENOSYLMETHIONINE, ... | | Authors: | Liu, X, Guo, D, Su, C, Chen, Y. | | Deposit date: | 2011-03-13 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex.
Plos Pathog., 7, 2011
|
|
7WD2
 
 | | Crystal structure of S43 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
7WD1
 
 | | Crystal structure of R14 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, R14, Spike protein S1, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
