2WNF
 
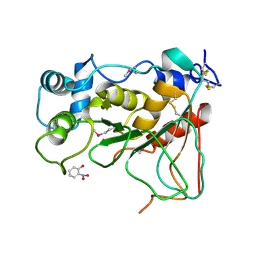 | | Crystal Structure of a Mammalian Sialyltransferase in complex with Gal-beta-1-3GalNAc-ortho-nitrophenol | | Descriptor: | CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, HYDROXY(2-HYDROXYPHENYL)OXOAMMONIUM, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Rao, F.V, Rich, J.R, Raikic, B, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-07-09 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Insight Into Mammalian Sialyltransferases.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1F00
 
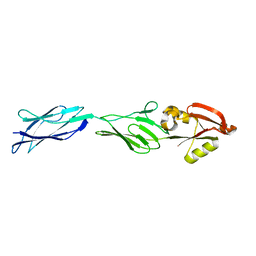 | | CRYSTAL STRUCTURE OF C-TERMINAL 282-RESIDUE FRAGMENT OF ENTEROPATHOGENIC E. COLI INTIMIN | | Descriptor: | INTIMIN | | Authors: | Luo, Y, Frey, E.A, Pfuetzner, R.A, Creagh, A.L, Knoechel, D.G, Haynes, C.A, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2000-05-12 | | Release date: | 2000-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex.
Nature, 405, 2000
|
|
2WML
 
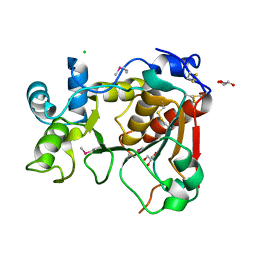 | | Crystal Structure of a Mammalian Sialyltransferase | | Descriptor: | CHLORIDE ION, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE -ALPHA-2,3-SIALYLTRANSFERASE, GLYCEROL | | Authors: | Rao, F.V, Rich, J.R, Raikic, B, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-07-01 | | Release date: | 2009-10-13 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight Into Mammalian Sialyltransferases.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2WQQ
 
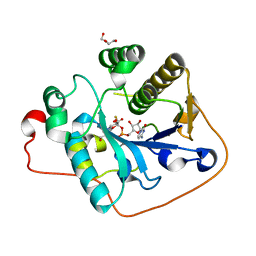 | | Crystallographic analysis of monomeric CstII | | Descriptor: | ALPHA-2,3-/2,8-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, DI(HYDROXYETHYL)ETHER | | Authors: | Chan, P.H.W, Lairson, L.L, Lee, H.J, Wakarchuk, W.W, Strynadka, N.C.J, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2009-08-25 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | NMR Spectroscopic Characterization of the Sialyltransferase Cstii from Camplyobacter Jejuni: Histidine 188 is the General Base.
Biochemistry, 48, 2009
|
|
1DLI
 
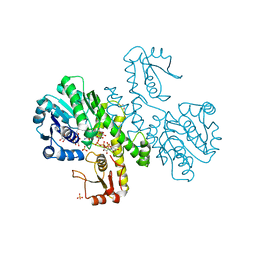 | | THE FIRST STRUCTURE OF UDP-GLUCOSE DEHYDROGENASE (UDPGDH) REVEALS THE CATALYTIC RESIDUES NECESSARY FOR THE TWO-FOLD OXIDATION | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Campbell, R.E, Mosimann, S.C, van de Rijn, I, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 1999-12-09 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
Biochemistry, 39, 2000
|
|
1DLJ
 
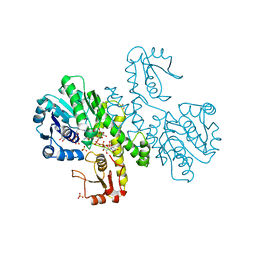 | | THE FIRST STRUCTURE OF UDP-GLUCOSE DEHYDROGENASE (UDPGDH) REVEALS THE CATALYTIC RESIDUES NECESSARY FOR THE TWO-FOLD OXIDATION | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Campbell, R.E, Mosimann, S.C, van de Rijn, I, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 1999-12-09 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
Biochemistry, 39, 2000
|
|
5TCQ
 
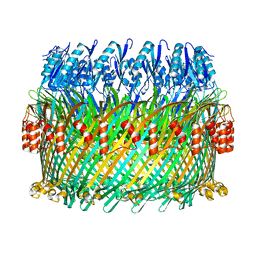 | | Near-atomic resolution cryo-EM structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG | | Descriptor: | Protein InvG | | Authors: | Worrall, L.J, Hong, C, Vuckovic, M, Bergeron, J.R.C, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2016-09-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body.
Nature, 540, 2016
|
|
5T91
 
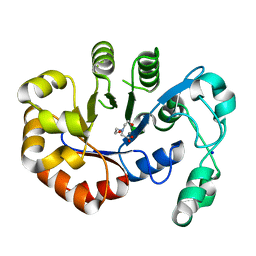 | | Crystal structure of B. subtilis 168 GlpQ in complex with bicine | | Descriptor: | BICINE, CALCIUM ION, Glycerophosphoryl diester phosphodiesterase, ... | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Identification of Two Phosphate Starvation-induced Wall Teichoic Acid Hydrolases Provides First Insights into the Degradative Pathway of a Key Bacterial Cell Wall Component.
J. Biol. Chem., 291, 2016
|
|
5TCR
 
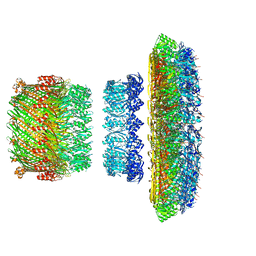 | | Atomic model of the Salmonella SPI-1 type III secretion injectisome basal body proteins InvG, PrgH, and PrgK | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH | | Authors: | Worrall, L.J, Hong, C, Vuckovic, M, Bergeron, J.R.C, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2016-09-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body.
Nature, 540, 2016
|
|
3GMW
 
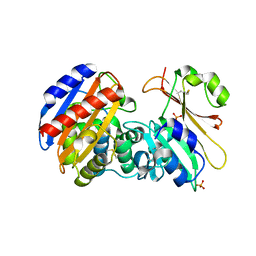 | | Crystal Structure of Beta-Lactamse Inhibitory Protein-I (BLIP-I) in Complex with TEM-1 Beta-Lactamase | | Descriptor: | B-lactamase, Beta-lactamase inhibitory protein BLIP-I, PHOSPHATE ION | | Authors: | Lim, D.C, Gretes, M, Strynadka, N.C.J. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
3GR5
 
 | |
3GR1
 
 | |
3GMY
 
 | |
8V33
 
 | |
8V34
 
 | |
8VA1
 
 | |
8VBT
 
 | |
3IR6
 
 | | Crystal structure of NarGHI mutant NarG-H49S | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bertero, M.G, Rothery, R.A, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2009-08-21 | | Release date: | 2010-01-05 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation.
J.Biol.Chem., 285, 2010
|
|
8VBU
 
 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Oxacillin) inhibited form | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBW
 
 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Ertapenem) inhibited form | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBV
 
 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Cephalexin) inhibited form | | Descriptor: | (2S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
1K0W
 
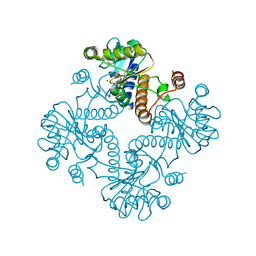 | | CRYSTAL STRUCTURE OF L-RIBULOSE-5-PHOSPHATE 4-EPIMERASE | | Descriptor: | L-RIBULOSE 5 PHOSPHATE 4-EPIMERASE, ZINC ION | | Authors: | Luo, Y, Samuel, J, Mosimann, S.C, Lee, J.E, Strynadka, N.C.J. | | Deposit date: | 2001-09-21 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of L-ribulose-5-phosphate 4-epimerase: an aldolase-like platform for epimerization
Biochemistry, 40, 2001
|
|
3IR5
 
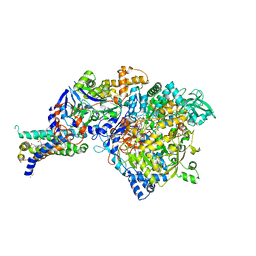 | | Crystal structure of NarGHI mutant NarG-H49C | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2009-08-21 | | Release date: | 2010-01-05 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation.
J.Biol.Chem., 285, 2010
|
|
5T9B
 
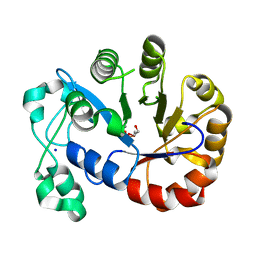 | |
3IR7
 
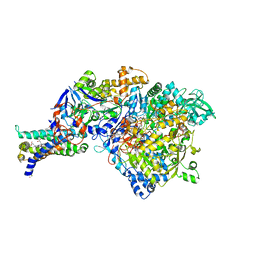 | | Crystal structure of NarGHI mutant NarG-R94S | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2009-08-21 | | Release date: | 2010-01-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation.
J.Biol.Chem., 285, 2010
|
|
