2WQQ
 
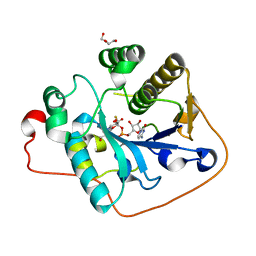 | | Crystallographic analysis of monomeric CstII | | Descriptor: | ALPHA-2,3-/2,8-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, DI(HYDROXYETHYL)ETHER | | Authors: | Chan, P.H.W, Lairson, L.L, Lee, H.J, Wakarchuk, W.W, Strynadka, N.C.J, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2009-08-25 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | NMR Spectroscopic Characterization of the Sialyltransferase Cstii from Camplyobacter Jejuni: Histidine 188 is the General Base.
Biochemistry, 48, 2009
|
|
2X4A
 
 | |
2YK7
 
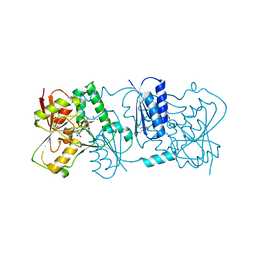 | | Structure of Neisseria LOS-specific sialyltransferase (NST), in complex with CMP-3F-Neu5Ac. | | Descriptor: | 1,2-ETHANEDIOL, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
2ZYL
 
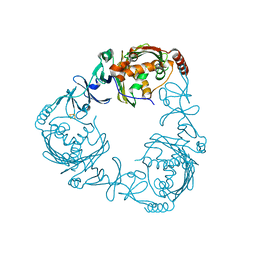 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase (KshA) from M. tuberculosis | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, POSSIBLE OXIDOREDUCTASE, ... | | Authors: | D'Angelo, I, Capyk, J, Strynadka, N, Eltis, L. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of 3-ketosteroid 9{alpha}-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis
J.Biol.Chem., 284, 2009
|
|
1XEB
 
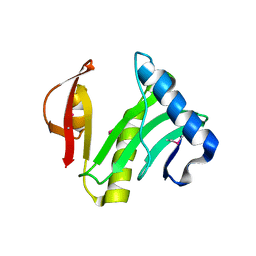 | | Crystal Structure of an Acyl-CoA N-acyltransferase from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein PA0115 | | Authors: | Bertero, M.G, Walker, J.R, Skarina, T, Gorodichtchenskaia, E, Joachimiak, A, Edwards, A.E, Savchenko, A, Strynadka, N, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-09 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of an Acyl-CoA N-acyltransferase from Pseudomonas aeruginosa
To be Published
|
|
2X49
 
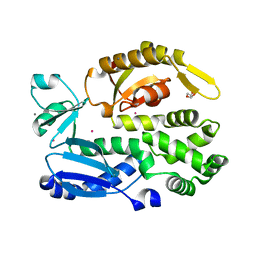 | | Crystal structure of the C-terminal domain of InvA | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, INVASION PROTEIN INVA, ... | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the C-Terminal Domain of the Salmonella Type III Secretion System Export Apparatus Protein Inva.
Protein Sci., 19, 2010
|
|
1XUU
 
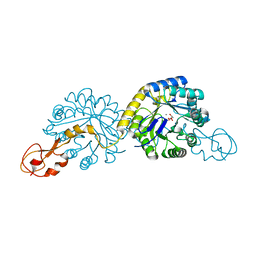 | | Crystal structure of sialic acid synthase (NeuB) in complex with Mn2+ and Malate from Neisseria meningitidis | | Descriptor: | D-MALATE, MANGANESE (II) ION, polysialic acid capsule biosynthesis protein SiaC | | Authors: | Gunawan, J, Simard, D, Gilbert, M, Lovering, A.L, Wakarchuk, W.W, Tanner, M.E, Strynadka, N.C. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol.
J.Biol.Chem., 280, 2005
|
|
1Y4Z
 
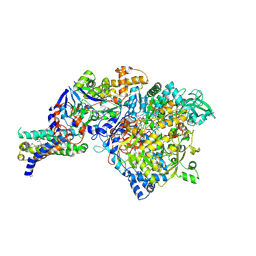 | | The crystal structure of Nitrate Reductase A, NarGHI, in complex with the Q-site inhibitor pentachlorophenol | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-01 | | Release date: | 2005-03-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1Y5L
 
 | | The crystal structure of the NarGHI mutant NarI-H66Y | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
2X62
 
 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE CST-II Y81F IN COMPLEX WITH CMP | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-2,3-/2,8-SIALYLTRANSFERASE, N3-PROTONATED CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
1XUZ
 
 | | Crystal structure analysis of sialic acid synthase (NeuB)from Neisseria meningitidis, bound to Mn2+, Phosphoenolpyruvate, and N-acetyl mannosaminitol | | Descriptor: | 5-DEOXY-5-{[(1S)-1-HYDROXYETHYL]AMINO}-D-GLUCITOL, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Gunawan, J, Simard, D, Gilbert, M, Lovering, A.L, Wakarchuk, W.W, Tanner, M.E, Strynadka, N.C. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol.
J.Biol.Chem., 280, 2005
|
|
1Y5N
 
 | | The crystal structure of the NarGHI mutant NarI-K86A in complex with pentachlorophenol | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1Y5I
 
 | | The crystal structure of the NarGHI mutant NarI-K86A | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
2X63
 
 | | Crystal structure of the sialyltransferase CST-II N51A in complex with CMP | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-2,3-/2,8-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
1XA1
 
 | | Crystal structure of the sensor domain of BlaR1 from Staphylococcus aureus in its apo form | | Descriptor: | PHOSPHATE ION, PYROPHOSPHATE 2-, Regulatory protein blaR1 | | Authors: | Wilke, M.S, Hills, T.L, Zhang, H.Z, Chambers, H.F, Strynadka, N.C. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus.
J.Biol.Chem., 279, 2004
|
|
1XSI
 
 | | Structure of a Family 31 alpha glycosidase | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ACETIC ACID, Putative family 31 glucosidase yicI, ... | | Authors: | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2004-10-19 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic and Structural Analysis of a Family 31 alpha-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
1XDY
 
 | | Structural and Biochemical Identification of a Novel Bacterial Oxidoreductase, W-containing cofactor | | Descriptor: | Bacterial Sulfite Oxidase, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, TUNGSTEN ION | | Authors: | Loschi, L, Brokx, S.J, Hills, T.L, Zhang, G, Bertero, M.G, Lovering, A.L, Weiner, J.H, Strynadka, N.C. | | Deposit date: | 2004-09-08 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical identification of a novel bacterial oxidoreductase.
J.Biol.Chem., 279, 2004
|
|
2P2V
 
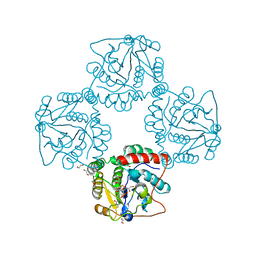 | | Crystal structure analysis of monofunctional alpha-2,3-sialyltransferase Cst-I from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, Alpha-2,3-sialyltransferase, CHLORIDE ION, ... | | Authors: | Chiu, C.P, Lairson, L.L, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2007-03-07 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of the alpha-2,3-Sialyltransferase Cst-I from Campylobacter jejuni in Apo and Substrate-Analogue Bound Forms.
Biochemistry, 46, 2007
|
|
2O4V
 
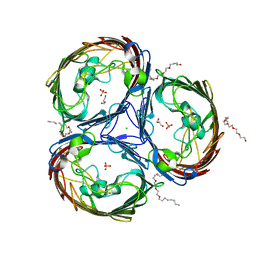 | | An arginine ladder in OprP mediates phosphate specific transfer across the outer membrane | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moraes, T.F, Bains, M, Hancock, R.E, Strynadka, N.C. | | Deposit date: | 2006-12-05 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | An arginine ladder in OprP mediates phosphate-specific transfer across the outer membrane.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1XDQ
 
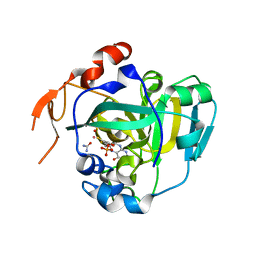 | | Structural and Biochemical Identification of a Novel Bacterial Oxidoreductase | | Descriptor: | Bacterial Sulfite Oxidase, MOLYBDENUM ATOM, OXYGEN ATOM, ... | | Authors: | Loschi, L, Brokx, S.J, Hills, T.L, Zhang, G, Bertero, M.G, Lovering, A.L, Weiner, J.H, Strynadka, N.C. | | Deposit date: | 2004-09-07 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and biochemical identification of a novel bacterial oxidoreductase.
J.Biol.Chem., 279, 2004
|
|
1XSJ
 
 | | Structure of a Family 31 alpha glycosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative family 31 glucosidase yicI | | Authors: | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2004-10-19 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and Structural Analysis of a Family 31 alpha-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
1Y9L
 
 | | The X-ray structure of an secretion system protein | | Descriptor: | ACETATE ION, Lipoprotein mxiM, UNDECANE | | Authors: | Lario, P.I, Pfuetzer, R.A, Frey, E.A, Creagh, L, Haynes, C, Maurelli, A.T, Strynadka, N.C. | | Deposit date: | 2004-12-15 | | Release date: | 2005-04-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and biochemical analysis of a secretin pilot protein.
Embo J., 24, 2005
|
|
1Y9T
 
 | |
2OLU
 
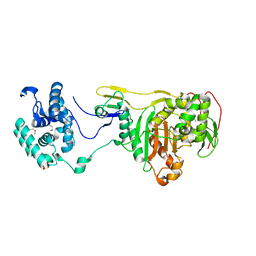 | | Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Penicillin-binding protein 2 | | Authors: | Lovering, A.L, De Castro, L.H, Lim, D, Strynadka, N.C. | | Deposit date: | 2007-01-19 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis.
Science, 315, 2007
|
|
1XSK
 
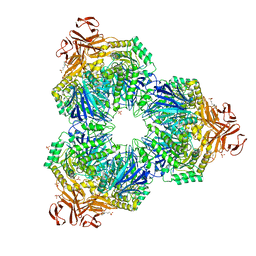 | | Structure of a Family 31 alpha glycosidase glycosyl-enzyme intermediate | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 5(R)-fluoro-beta-D-xylopyranose, Putative family 31 glucosidase yicI, ... | | Authors: | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2004-10-19 | | Release date: | 2004-11-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic and Structural Analysis of a Family 31 {alpha}-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
