1YLI
 
 | |
1HTW
 
 | |
4K2A
 
 | | Crystal structure of haloalkane dehalogenase DbeA from Bradyrhizobium elkani USDA94 | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Prudnikova, T, Chaloupkova, R, Rezacova, P, Mozga, T, Koudelakova, T, Sato, Y, Kuty, M, Nagata, Y, Damborsky, J, Kuta Smatanova, I, Structure 2 Function Project (S2F) | | Deposit date: | 2013-04-08 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of a novel haloalkane dehalogenase with two halide-binding sites.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2OUT
 
 | | Solution Structure of HI1506, a Novel Two Domain Protein from Haemophilus influenzae | | Descriptor: | Mu-like prophage FluMu protein gp35, Protein HI1507 in Mu-like prophage FluMu region | | Authors: | Sari, N, He, Y, Doseeva, V, Surabian, K, Schwarz, F, Herzberg, O, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2007-02-12 | | Release date: | 2007-05-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HI1506, a novel two-domain protein from Haemophilus influenzae.
Protein Sci., 16, 2007
|
|
3PB1
 
 | | Crystal Structure of a Michaelis Complex between Plasminogen Activator Inhibitor-1 and Urokinase-type Plasminogen Activator | | Descriptor: | Plasminogen activator inhibitor 1, Plasminogen activator, urokinase, ... | | Authors: | Lin, Z, Jiang, L, Huang, M, Structure 2 Function Project (S2F) | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of urokinase-type plasminogen activator by plasminogen activator inhibitor-1.
J.Biol.Chem., 286, 2011
|
|
3BJK
 
 | | Crystal structure of HI0827, a hexameric broad specificity acyl-coenzyme A thioesterase: The Asp44Ala mutant enzyme | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA thioester hydrolase HI0827, CITRIC ACID | | Authors: | Willis, M.A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2007-12-04 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of YciA from Haemophilus influenzae (HI0827), a Hexameric Broad Specificity Acyl-Coenzyme A Thioesterase.
Biochemistry, 47, 2008
|
|
3CA8
 
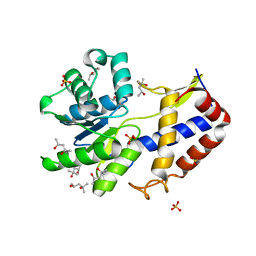 | | Crystal structure of Escherichia coli YdcF, an S-adenosyl-L-methionine utilizing enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Protein ydcF, SULFATE ION | | Authors: | Lim, K, Chao, K, Lehmann, C, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2008-02-19 | | Release date: | 2008-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Escherichia coli YdcF binds S-adenosyl-L-methionine and adopts an alpha/beta-fold characteristic of nucleotide-utilizing enzymes.
Proteins, 72, 2008
|
|
1DBU
 
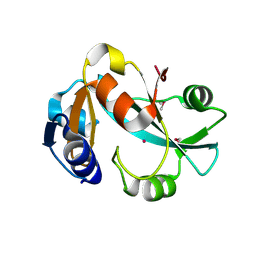 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase protein from H. influenzae (HI1434) | | Descriptor: | MERCURY (II) ION, cysteinyl-tRNA(Pro) deacylase | | Authors: | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
1FL9
 
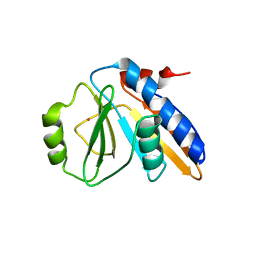 | | THE YJEE PROTEIN | | Descriptor: | HYPOTHETICAL PROTEIN HI0065, MERCURY (II) ION | | Authors: | Teplyakov, A, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2000-08-13 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the YjeE protein from Haemophilus influenzae: a putative Atpase involved in cell wall synthesis
Proteins, 48, 2002
|
|
1DBX
 
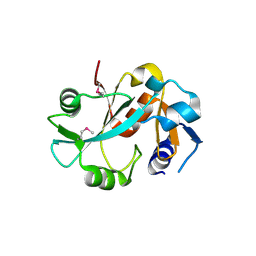 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase from H. influenzae (HI1434) | | Descriptor: | cysteinyl-tRNA(Pro) deacylase | | Authors: | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
1XAX
 
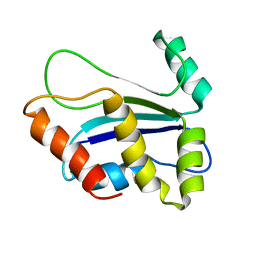 | | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae | | Descriptor: | Hypothetical UPF0054 protein HI0004 | | Authors: | Yeh, D.C, Parsons, J.F, Parsons, L.M, Liu, F, Eisenstein, E, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-26 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae, and comparison with the X-ray structure of an Aquifex aeolicus homolog
Protein Sci., 14, 2005
|
|
1J8B
 
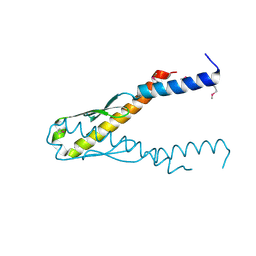 | | Structure of YbaB from Haemophilus influenzae (HI0442), a protein of unknown function | | Descriptor: | YbaB | | Authors: | Lim, K, Tempcyzk, A, Toedt, J, Parsons, J.F, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-21 | | Release date: | 2003-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of YbaB from Haemophilus influenzae (HI0442), a
protein of unknown function coexpressed with the recombinational
DNA repair protein RecR
Proteins, 50, 2003
|
|
1IMU
 
 | | Solution Structure of HI0257, a Ribosome Binding Protein | | Descriptor: | HYPOTHETICAL PROTEIN HI0257 | | Authors: | Parsons, L, Eisenstein, E, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-11 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HI0257, a bacterial ribosome binding protein.
Biochemistry, 40, 2001
|
|
1IN0
 
 | |
1IZM
 
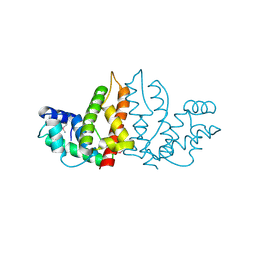 | | Structure of ygfB from Haemophilus influenzae (HI0817), a Conserved Hypothetical Protein | | Descriptor: | HYPOTHETICAL PROTEIN HI0817 | | Authors: | Galkin, A, Sarikaya, E, Lehmann, C, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-10-09 | | Release date: | 2003-12-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray structure of HI0817 from Haemophilus influenzae: protein of unknown function with a novel fold.
Proteins, 57, 2004
|
|
1JOS
 
 | | Ribosome Binding Factor A(rbfA) | | Descriptor: | RIBOSOME-BINDING FACTOR A | | Authors: | Bonander, N, Tordova, M, Howard, A.J, Eisenstein, E, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-30 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7-A Crystal Structure of HI1288 - Ribosome Binding Factor A (rbfA), a Cold Response Protein
To be Published
|
|
1JOG
 
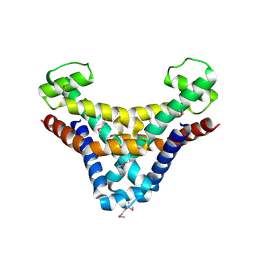 | | Structure of HI0074 from Heamophilus Influenzae reveals the fold of a substrate binding domain of a nucleotidyltransferase | | Descriptor: | HYPOTHETICAL PROTEIN HI0074 | | Authors: | Lehmann, C, Lim, K, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-29 | | Release date: | 2002-12-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The HI0073/HI0074 protein pair from Haemophilus influenzae is a member of a new nucleotidyltransferase family: Structure, sequence analyses, and
solution studies
Proteins, 50, 2003
|
|
1NNV
 
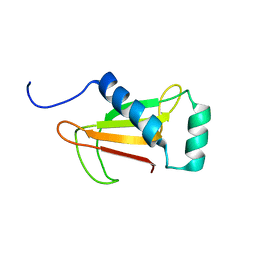 | |
1JAL
 
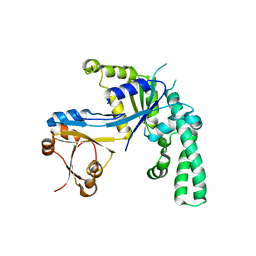 | |
1LQA
 
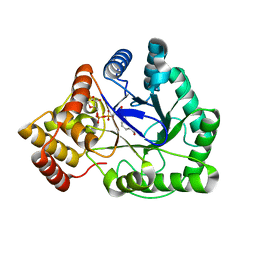 | | TAS PROTEIN FROM ESCHERICHIA COLI IN COMPLEX WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Tas protein | | Authors: | Obmolova, G, Teplyakov, A, Khil, P.P, Howard, A.J, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2002-05-09 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Crystal structure of the Escherichia coli Tas protein, an NADP(H)-dependent aldo-keto reductase
PROTEINS: STRUCT.,FUNCT.,GENET., 53, 2003
|
|
1J7H
 
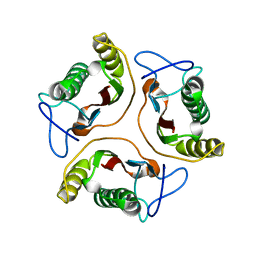 | | Solution Structure of HI0719, a Hypothetical Protein From Haemophilus Influenzae | | Descriptor: | HYPOTHETICAL PROTEIN HI0719 | | Authors: | Parsons, L, Bonander, N, Eisenstein, E, Gilson, M, Kairys, V, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-16 | | Release date: | 2003-02-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Functional Ligand Screening of HI0719, a Highly Conserved Protein from Bacteria to Humans in the YjgF/YER057c/UK114 Family
Biochemistry, 42, 2003
|
|
1J7G
 
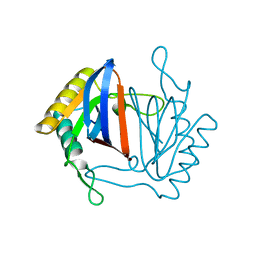 | |
1IM8
 
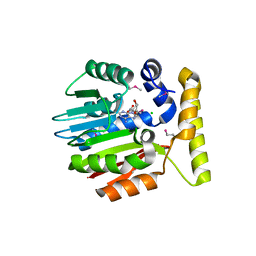 | | Crystal structure of YecO from Haemophilus influenzae (HI0319), a methyltransferase with a bound S-adenosylhomocysteine | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOSELENOCYSTEINE, YecO | | Authors: | Lim, K, Zhang, H, Tempczyk, A, Bonander, N, Toedt, J, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-10 | | Release date: | 2001-11-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of YecO from Haemophilus influenzae (HI0319) reveals a methyltransferase fold and a bound S-adenosylhomocysteine.
Proteins, 45, 2001
|
|
1JOP
 
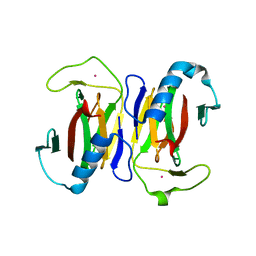 | |
1JO0
 
 | | Structure of HI1333, a Hypothetical Protein from Haemophilus influenzae with Structural Similarity to RNA-binding Proteins | | Descriptor: | GLYCEROL, HYPOTHETICAL PROTEIN HI1333 | | Authors: | Willis, M.A, Krajewski, W, Chalamasetty, V.R, Reddy, P, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-26 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of HI1333 (YhbY), a putative RNA-binding protein from Haemophilus influenzae
Proteins, 49, 2002
|
|
