2YWY
 
 | |
2Z8W
 
 | | Structure of an IgNAR-AMA1 complex | | Descriptor: | Apical membrane antigen 1, New antigen receptor variable domain | | Authors: | Streltsov, V.A, Henderson, K.A, Batchelor, A.H, Coley, A.M, Nuttall, S.D. | | Deposit date: | 2007-09-11 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of an IgNAR-AMA1 Complex: Targeting a Conserved Hydrophobic Cleft Broadens Malarial Strain Recognition
Structure, 15, 2007
|
|
3AAZ
 
 | |
3W09
 
 | | Influenza virus neuraminidase subtype N9 (TERN) complexed with 2,3-dif guanidino-neu5ac2en inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-3-fluoro-D-glycero-D-galacto-non-2-enonic acid, ... | | Authors: | Streltsov, V.A. | | Deposit date: | 2012-10-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism-based covalent neuraminidase inhibitors with broad-spectrum influenza antiviral activity
Science, 340, 2013
|
|
3WLR
 
 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL | | Authors: | Streltsov, V.A, Hrmova, M, Luang, S. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLH
 
 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLQ
 
 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL | | Authors: | Streltsov, V.A, Luang, S, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLJ
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with 3-deoxy-glucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLL
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with PEG400 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[beta-D-xylopyranose-(1-2)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLN
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with octyl-S-glucoside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLK
 
 | |
3WLM
 
 | |
3WLO
 
 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, SULFATE ION, ... | | Authors: | Streltsov, V.A, Luang, S, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLP
 
 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 1-thio-beta-D-glucopyranose-(1-6)-methyl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, ... | | Authors: | Streltsov, V.A, Luang, S, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLI
 
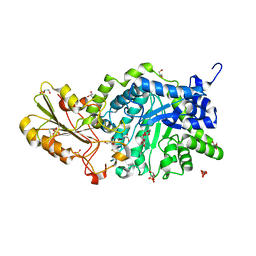 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
4F37
 
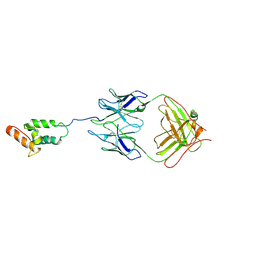 | | Structure of the tethered N-terminus of Alzheimer's disease A peptide | | Descriptor: | Colicin-E7 immunity protein, Fab WO2 anti-amyloid-beta antibody Fab fragment, Im7 immunity protein | | Authors: | Nisbet, R.M, Nuttall, S.D, Caine, J.M, Rober, R, Hittaki, M, Pearce, L.A, Davydova, N, Masters, C.L, Varghese, J.N, Streltsov, V.A. | | Deposit date: | 2012-05-09 | | Release date: | 2013-05-15 | | Last modified: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural studies of the tethered N-terminus of the Alzheimer's disease amyloid-beta peptide.
Proteins, 81, 2013
|
|
6LBB
 
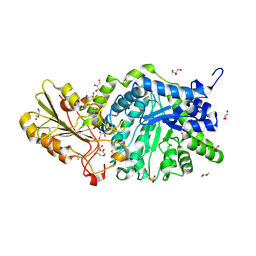 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-D-glucan exohydrolase isoenzyme ExoI, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6LBV
 
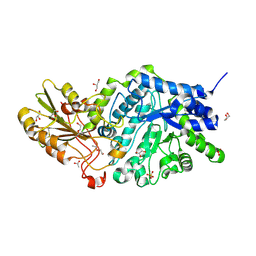 | | Crystal structure of barley exohydrolaseI W434F mutant in complex with methyl 6-thio-beta-gentiobioside | | Descriptor: | ACETATE ION, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-11-15 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
4OZT
 
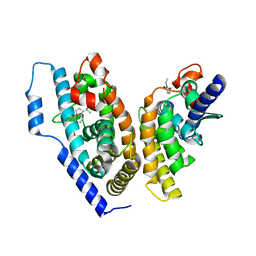 | | crystal structure of the ligand binding domains of the Bovicola ovis ecdysone receptor EcR/USP heterodimer (PonA crystal) | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor, N-ETHYLMALEIMIDE, ... | | Authors: | Ren, B, Peat, T.S, Streltsov, V.A, Pollard, M, Fernley, R, Grusovin, J, Seabrook, S, Pilling, P, Phan, T, Lu, L, Lovrecz, G.O, Graham, L.D, Hill, R.J. | | Deposit date: | 2014-02-19 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unprecedented conformational flexibility revealed in the ligand-binding domains of the Bovicola ovis ecdysone receptor (EcR) and ultraspiracle (USP) subunits.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OZR
 
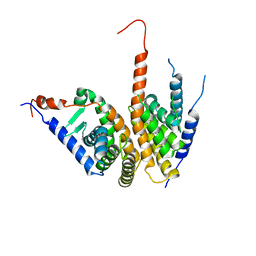 | | Crystal structure of the ligand binding domains of the Bovicola ovis ecdysone receptor EcR/USP heterodimer (methylene lactam crystal) | | Descriptor: | Ecdysone receptor, Retinoid X receptor | | Authors: | Ren, B, Peat, T.S, Streltsov, V.A, Pollard, M, Fernley, R, Grusovin, J, Seabrook, S, Pilling, P, Phan, T, Lu, L, Lovrecz, G.O, Graham, L.D, Hill, R.J. | | Deposit date: | 2014-02-18 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unprecedented conformational flexibility revealed in the ligand-binding domains of the Bovicola ovis ecdysone receptor (EcR) and ultraspiracle (USP) subunits.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1I4J
 
 | | CRYSTAL STRUCTURE OF L22 RIBOSOMAL PROTEIN MUTANT | | Descriptor: | 50S RIBOSOMAL PROTEIN L22 | | Authors: | Davydova, N.L, Streltsov, V.A, Fedorov, R, Wilce, M, Liljas, A, Garder, M. | | Deposit date: | 2001-02-22 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | L22 ribosomal protein and effect of its mutation on ribosome resistance to erythromycin.
J.Mol.Biol., 322, 2002
|
|
1X39
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole | | Descriptor: | (5R,6R,7S,8S)-3-(ANILINOMETHYL)-5,6,7,8-TETRAHYDRO-5-(HYDROXYMETHYL)-IMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[beta-D-xylopyranose-(1-2)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Hrmova, M, Streltsov, V.A, Smith, B.J, Vasella, A, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2005-05-02 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural rationale for low-nanomolar binding of transition state mimics to a family GH3 beta-D-glucan glucohydrolase from barley.
Biochemistry, 44, 2005
|
|
1X38
 
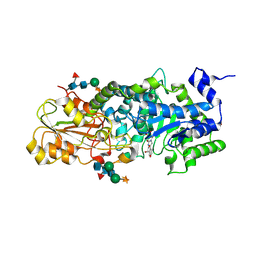 | | crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole | | Descriptor: | (5R,6R,7S,8S)-5-(HYDROXYMETHYL)-2-PHENYL-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[beta-D-xylopyranose-(1-2)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Hrmova, M, Streltsov, V.A, Smith, B.J, Vasella, A, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2005-05-02 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structural rationale for low-nanomolar binding of transition state mimics to a family GH3 beta-D-glucan glucohydrolase from barley.
Biochemistry, 44, 2005
|
|
1V2I
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
1V3B
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
