7QLG
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT L259V COMPLEXED WITH FE, NADH, AND GLYCEROL | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE (III) ION, GLYCEROL, ... | | Authors: | Sridhar, S, Kiema, T.R, Widertsen, M, Wierenga, R.K. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and kinetic studies of a laboratory evolved aldehyde reductase explain the dramatic shift of its new substrate specificity.
Iucrj, 10, 2023
|
|
7QLS
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT N151G, L259V COMPLEXED WITH FE, NADH, AND DIMETHOXYPHENYL ACETAMIDE | | Descriptor: | 2-(3,4-dimethoxyphenyl)ethanamide, ADENOSINE-5-DIPHOSPHORIBOSE, FE (III) ION, ... | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R.K, Widersten, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and kinetic studies of a laboratory evolved aldehyde reductase explain the dramatic shift of its new substrate specificity.
Iucrj, 10, 2023
|
|
7QNH
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT N151G, L259V COMPLEXED WITH FE, NADH, AND GLYCEROL | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE (III) ION, GLYCEROL, ... | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R.K, Widersten, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and kinetic studies of a laboratory evolved aldehyde reductase explain the dramatic shift of its new substrate specificity.
Iucrj, 10, 2023
|
|
7QLQ
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT N151G, L259V COMPLEXED WITH FE, NAD, AND DIMETHOXYPHENYL ACETAMIDE | | Descriptor: | 2-(3,4-dimethoxyphenyl)ethanamide, ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, ... | | Authors: | Sridhar, S, Kiema, T.R, Widersten, M, Wierenga, R.K. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures and kinetic studies of a laboratory evolved aldehyde reductase explain the dramatic shift of its new substrate specificity.
Iucrj, 10, 2023
|
|
7R5T
 
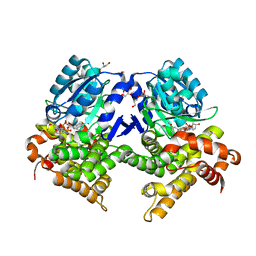 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT F254I COMPLEXED WITH FE, NADH, AND GLYCEROL | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE-5-DIPHOSPHORIBOSE, FE (III) ION, ... | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R, Widersten, M. | | Deposit date: | 2022-02-11 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
7R3D
 
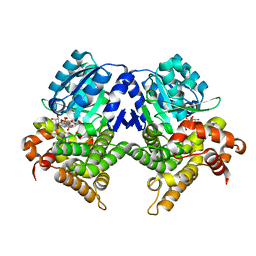 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT N151G, L259V COMPLEXED WITH FE, NADH, AND GLYCEROL (Absence of Nicotinamide ring) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, FE (III) ION, Lactaldehyde reductase | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R, Widersten, M. | | Deposit date: | 2022-02-07 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
7QNF
 
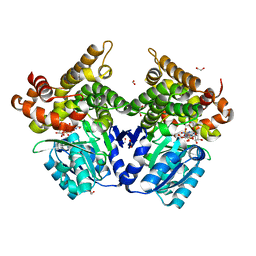 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT N151G, L259V COMPLEXED WITH FE, NAD+, AND ETHYLENE GLYCOL | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, FE (III) ION, ... | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R.K, Widersten, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
7QNI
 
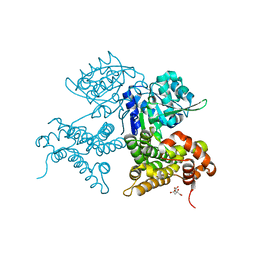 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT L259V | | Descriptor: | CITRIC ACID, Lactaldehyde reductase | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R.K, Widersten, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
7QNJ
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT F254I COMPLEXED WITH FE, NAD+, AND GLYCEROL | | Descriptor: | FE (III) ION, GLYCEROL, Lactaldehyde reductase, ... | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R.K, Widersten, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
3HWO
 
 | |
2GP6
 
 | |
1Y50
 
 | | X-ray crystal structure of Bacillus stearothermophilus Histidine phosphocarrier protein (Hpr) F29W mutant domain_swapped dimer | | Descriptor: | Phosphocarrier protein HPr, SULFATE ION | | Authors: | Sridharan, S, Razvi, A, Scholtz, J.M, Sacchettini, J.C. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The HPr proteins from the thermophile Bacillus stearothermophilus can form domain-swapped dimers.
J.Mol.Biol., 346, 2005
|
|
1Y4Y
 
 | | X-ray crystal structure of Bacillus stearothermophilus Histidine phosphocarrier protein (Hpr) | | Descriptor: | Phosphocarrier protein HPr, SULFATE ION | | Authors: | Sridharan, S, Razvi, A, Scholtz, J.M, Sacchettini, J.C. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The HPr proteins from the thermophile Bacillus stearothermophilus can form domain-swapped dimers.
J.Mol.Biol., 346, 2005
|
|
1Y51
 
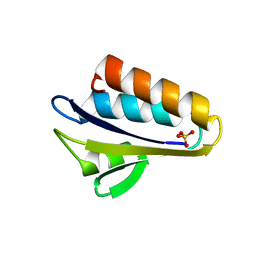 | | X-ray crystal structure of Bacillus stearothermophilus Histidine phosphocarrier protein (Hpr) F29W mutant | | Descriptor: | Phosphocarrier protein HPr, SULFATE ION | | Authors: | Sridharan, S, Razvi, A, Scholtz, J.M, Sacchettini, J.C. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The HPr proteins from the thermophile Bacillus stearothermophilus can form domain-swapped dimers.
J.Mol.Biol., 346, 2005
|
|
6Z5F
 
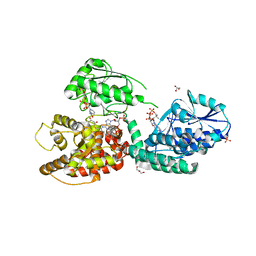 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE-1 (RPMFE1) COMPLEXED WITH 3-KETODECANOYL-COA AND OXIDISED NICOTINAMIDE ADENINE DINUCLEOTIDE | | Descriptor: | 3-KETO-DECANOYL-COA, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Wierenga, R.K, Sridhar, S, Kiema, T.R. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic binding studies of rat peroxisomal multifunctional enzyme type 1 with 3-ketodecanoyl-CoA: capturing active and inactive states of its hydratase and dehydrogenase catalytic sites.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZIB
 
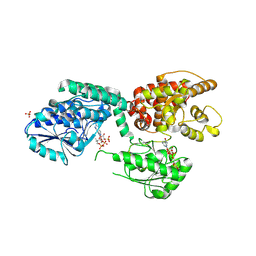 | |
6ZIC
 
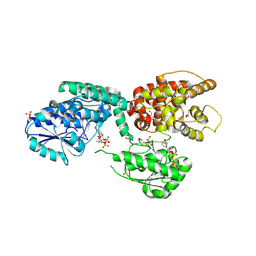 | |
6Z5V
 
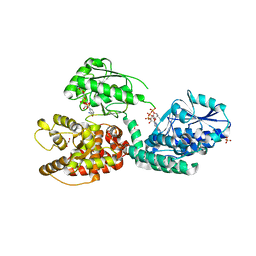 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE-1 (RPMFE1) COMPLEXED WITH 3-KETODECANOYL-COA IN CROTONASE FOLD AND OXIDISED NICOTINAMIDE ADENINE DINUCLEOTIDE IN HAD FOLD | | Descriptor: | 3-KETO-DECANOYL-COA, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Wierenga, R.K, Sridhar, S, Kiema, T.R. | | Deposit date: | 2020-05-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystallographic binding studies of rat peroxisomal multifunctional enzyme type 1 with 3-ketodecanoyl-CoA: capturing active and inactive states of its hydratase and dehydrogenase catalytic sites.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6Z5O
 
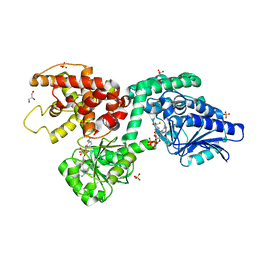 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE-1 (RPMFE1) COMPLEXED WITH COENZYME-A AND OXIDISED NICOTINAMIDE ADENINE DINUCLEOTIDE | | Descriptor: | COENZYME A, GLYCEROL, NICOTINAMIDE, ... | | Authors: | Wierenga, R.K, Sridhar, S, Kiema, T.R. | | Deposit date: | 2020-05-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic binding studies of rat peroxisomal multifunctional enzyme type 1 with 3-ketodecanoyl-CoA: capturing active and inactive states of its hydratase and dehydrogenase catalytic sites.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4UV4
 
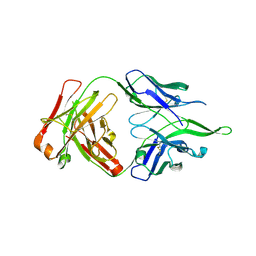 | | Crystal structure of anti-FPR Fpro0165 Fab fragment | | Descriptor: | FPRO0165 FAB | | Authors: | Douthwaite, J.A, Sridharan, S, Huntington, C, Marwood, R, Hammersley, J, Hakulinen, J.K, Ek, M, Sjogren, T, Rider, D, Privezentzev, C, Seaman, J.C, Cariuk, P, Knights, V, Young, J, Wilkinson, T, Sleeman, M, Finch, D.K, Lowe, D.C, Vaughan, T.J. | | Deposit date: | 2014-08-04 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Affinity Maturation of a Novel Antagonistic Human Monoclonal Antibody with a Long Vh Cdr3 Targeting the Class a Gpcr Formyl-Peptide Receptor 1.
Mabs, 7, 2015
|
|
4XGM
 
 | |
4XGL
 
 | |
4UU9
 
 | | Crystal structure of the human c5a in complex with MEDI7814 a neutralising antibody | | Descriptor: | COMPLEMENT C5, MEDI7814, SULFATE ION | | Authors: | Colley, C, Sridharan, S, Dobson, C, Popovic, B, Debreczeni, J.E, Hargreaves, D, Edwards, B, Brennan, J, England, L, Fung, S, An Eghobamien, L, Sivars, U, Woods, R, Flavell, L, Renshaw, G.J, Wickson, K, Wilkinson, T, Davies, R, Bonnell, J, Warrener, P, Howes, R, Vaughan, T. | | Deposit date: | 2014-07-25 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and characterization of a high affinity C5a monoclonal antibody that blocks binding to C5aR1 and C5aR2 receptors.
MAbs, 10, 2018
|
|
6XN4
 
 | | Structure of the Lactococcus lactis Csm CTR_3:2 CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm2, CRISPR-associated protein Csm3, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
6XN5
 
 | | Structure of the Lactococcus lactis Csm Apo- CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm3, CRISPR-associated protein Csm4, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
