8FC0
 
 | |
8T9W
 
 | |
7T7Y
 
 | |
7T7Z
 
 | | The crystal structure of family 8 carbohydrate-binding module from Dictyostelium discoideum | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, Endoglucanase, ... | | Authors: | Marcelo, M.V, Campos, B.M, Squina, F.M. | | Deposit date: | 2021-12-15 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Unique properties of a Dictyostelium discoideum carbohydrate-binding module expand our understanding of CBM-ligand interactions.
J.Biol.Chem., 298, 2022
|
|
6P5U
 
 | | Structure of an enoyl-CoA hydratase/aldolase isolated from a lignin-degrading consortium | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COENZYME A, Enoyl-CoA hydratase | | Authors: | Liberato, M.V, Squina, F.M. | | Deposit date: | 2019-05-31 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a prokaryotic feruloyl-CoA hydratase-lyase from a lignin-degrading consortium with high oligomerization stability under extreme pHs.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
5KLF
 
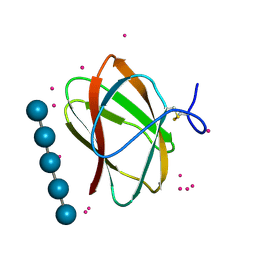 | | Structure of CBM_E1, a novel carbohydrate-binding module found by sugar cane soil metagenome, complexed with cellopentaose and gadolinium ion | | Descriptor: | Carbohydrate binding module E1, GADOLINIUM ATOM, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Liberato, M.V, Campos, B.M, Zeri, A.C.M, Squina, F.M. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | A Novel Carbohydrate-binding Module from Sugar Cane Soil Metagenome Featuring Unique Structural and Carbohydrate Affinity Properties.
J.Biol.Chem., 291, 2016
|
|
5KLE
 
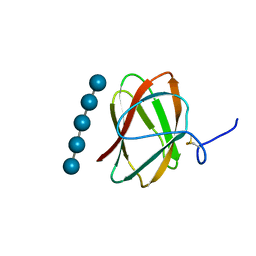 | | Structure of CBM_E1, a novel carbohydrate-binding module found by sugar cane soil metagenome, complexed with cellopentaose | | Descriptor: | Carbohydrate binding module E1, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Liberato, M.V, Campos, B.M, Zeri, A.C.M, Squina, F.M. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Novel Carbohydrate-binding Module from Sugar Cane Soil Metagenome Featuring Unique Structural and Carbohydrate Affinity Properties.
J.Biol.Chem., 291, 2016
|
|
5KLC
 
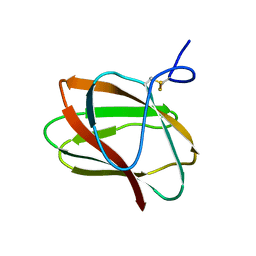 | | Structure of CBM_E1, a novel carbohydrate-binding module found by sugar cane soil metagenome | | Descriptor: | Carbohydrate binding module E1 | | Authors: | Liberato, M.V, Campos, B.M, Zeri, A.C.M, Squina, F.M. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | A Novel Carbohydrate-binding Module from Sugar Cane Soil Metagenome Featuring Unique Structural and Carbohydrate Affinity Properties.
J.Biol.Chem., 291, 2016
|
|
5KET
 
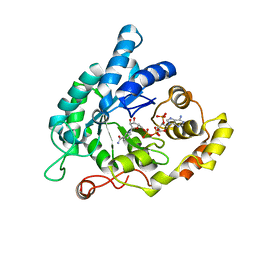 | | Structure of the aldo-keto reductase from Coptotermes gestroi | | Descriptor: | Aldo-keto reductase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liberato, M.L, Campos, B.M, Tramontina, R, Squina, F.M. | | Deposit date: | 2016-06-10 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Coptotermes gestroi aldo-keto reductase: a multipurpose enzyme for biorefinery applications.
Biotechnol Biofuels, 10, 2017
|
|
6TN6
 
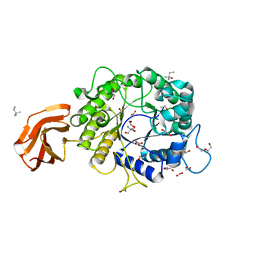 | | X-ray structure of the endo-beta-1,4-mannanase from Thermotoga petrophila | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | da Silva, V.M, Squina, F.M, Sperenca, M, Martin, L, Muniz, J.R.C, Garcia, W, Nicolet, Y. | | Deposit date: | 2019-12-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution structure of a modular hyperthermostable endo-beta-1,4-mannanase from Thermotoga petrophila: The ancillary immunoglobulin-like module is a thermostabilizing domain.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
5UBJ
 
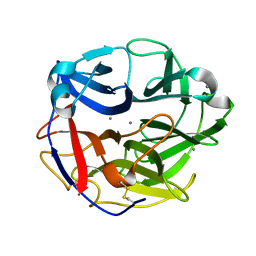 | | Structure of an alpha-L-arabinofuranosidase (GH62) from Aspergillus nidulans | | Descriptor: | Alpha-L-arabinofuranosidase axhA-2, CALCIUM ION | | Authors: | Liberato, M.V, Contesini, F.J, Damasio, A.R, Squina, F.M. | | Deposit date: | 2016-12-20 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of a highly secreted alpha-l-arabinofuranosidase (GH62) from Aspergillus nidulans grown on sugarcane bagasse.
Biochim. Biophys. Acta, 1865, 2017
|
|
4DFS
 
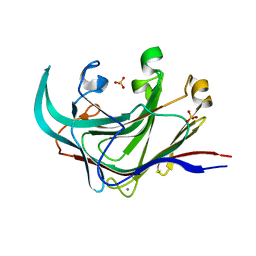 | | Structure of the catalytic domain of an endo-1,3-beta-glucanase (laminarinase) from Thermotoga petrophila RKU-1 | | Descriptor: | CALCIUM ION, Glycoside hydrolase, family 16, ... | | Authors: | Meza, A.N, Ruller, R, Prade, R.A, Squina, F.M, Santos, C.R, Murakami, M.T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.754 Å) | | Cite: | Structural studies of an endo-1,3-beta-glucanase from Thermotoga petrophila RKU-1
To be Published
|
|
2YOK
 
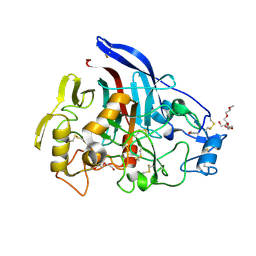 | | Cellobiohydrolase I Cel7A from Trichoderma harzianum at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Textor, L.C, Colussi, F, Serpa, V, Squina, F.M, Pereira Jr, N, Polikarpov, I. | | Deposit date: | 2012-10-25 | | Release date: | 2012-11-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Joint X-Ray Crystallographic and Molecular Dynamics Study of Cellobiohydrolase I from Trichoderma Harzianum: Deciphering the Structural Features of Cellobiohydrolase Catalytic Activity.
FEBS J., 280, 2013
|
|
2Y9N
 
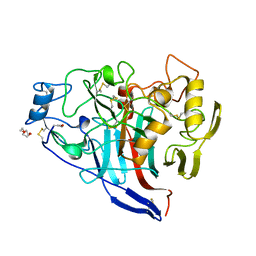 | | Cellobiohydrolase I Cel7A from Trichoderma harzianum at 2.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EXOGLUCANASE 1, TRIETHYLENE GLYCOL | | Authors: | Textor, L.C, Colussi, F, Serpa, V, Squina, F, Pereira Jr, N, Polikarpov, I. | | Deposit date: | 2011-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Cellobiohydrolase I from Trichoderma Harzianum: Structural and Enzymatic Characterization
To be Published
|
|
6XQM
 
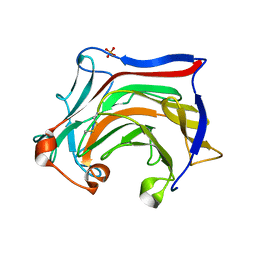 | | Crystal structure of SCLam E144S mutant, a non-specific endo-beta-1,3(4)-glucanase from family GH16, co-crystallized with laminarihexaose, presenting a laminaribiose and a glucose at active site | | Descriptor: | CALCIUM ION, GH16 family protein, PHOSPHATE ION, ... | | Authors: | Liberato, M.V, Squina, F. | | Deposit date: | 2020-07-09 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
6XOF
 
 | | Crystal structure of SCLam, a non-specific endo-beta-1,3(4)-glucanase from family GH16 | | Descriptor: | CALCIUM ION, GH16 family protein, GLYCEROL | | Authors: | Liberato, M.V, Bernardes, A, Polikarpov, I, Squina, F. | | Deposit date: | 2020-07-07 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
6XQL
 
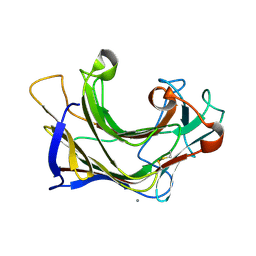 | | Crystal structure of SCLam E144S mutant, a non-specific endo-beta-1,3(4)-glucanase from family GH16, co-crystallized with cellohexaose, presenting a 1,3-beta-D-cellobiosyl-glucose at active site | | Descriptor: | CALCIUM ION, GH16 family protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Liberato, M.V, Squina, F. | | Deposit date: | 2020-07-09 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
6XQF
 
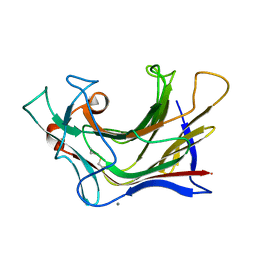 | | Crystal structure of SCLam E144S mutant, a non-specific endo-beta-1,3(4)-glucanase from family GH16, co-crystallized with 1,3-beta-D-cellotriosyl-glucose, presenting a 1,3-beta-D-cellobiosyl-glucose at active site | | Descriptor: | CALCIUM ION, GH16 family protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Liberato, M.V, Squina, F. | | Deposit date: | 2020-07-09 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
6XQH
 
 | | Crystal structure of SCLam E144S mutant, a non-specific endo-beta-1,3(4)-glucanase from family GH16, co-crystallized with cellotriose, presenting a 1,3-beta-D-cellobiosyl-glucose and a cellobiose at active site | | Descriptor: | CALCIUM ION, GH16 family protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Liberato, M.V, Squina, F. | | Deposit date: | 2020-07-09 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
6XQG
 
 | | Crystal structure of SCLam E144S mutant, a non-specific endo-beta-1,3(4)-glucanase from family GH16, co-crystallized with 1,3-beta-D-cellobiosyl-cellobiose, presenting a 1,3-beta-D-cellobiosyl-glucose at active site | | Descriptor: | CALCIUM ION, GH16 family protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Liberato, M.V, Squina, F. | | Deposit date: | 2020-07-09 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
3PZV
 
 | | C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 | | Descriptor: | Endoglucanase | | Authors: | Santos, C.R, Paiva, J.H, Akao, P.K, Meza, A.N, Silva, J.C, Squina, F.M, Ward, R.J, Ruller, R, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.867 Å) | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
4K68
 
 | | Structure of a novel GH10 endoxylanase retrieved from sugarcane soil metagenome | | Descriptor: | GH10 xylanase, GLYCEROL | | Authors: | Santos, C.R, Polo, C.C, Alvarez, T.M, Paixao, D.A.A, Almeida, R.F, Pereira, I.O, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-04-15 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Development and biotechnological application of a novel endoxylanase family GH10 identified from sugarcane soil metagenome.
Plos One, 8, 2013
|
|
3PZT
 
 | | Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion | | Descriptor: | Endoglucanase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Santos, C.R, Paiva, J.H, Akao, P.K, Meza, A.N, Silva, J.C, Squina, F.M, Ward, R.J, Ruller, R, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
4KC7
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Glycoside hydrolase, ... | | Authors: | Nascimento, A.F.Z, Polo, C.C, Santos, C.R, Costa, M.C.M.F, Mesa, A.N, Prade, R.A, Ruller, R, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
3S2C
 
 | | Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 | | Descriptor: | Alpha-N-arabinofuranosidase | | Authors: | Souza, T.A.C.B, Santos, C.R, Souza, A.R, Oldiges, D.P, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2011-05-16 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a novel thermostable GH51 Alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1
Protein Sci., 20, 2011
|
|
