5VIJ
 
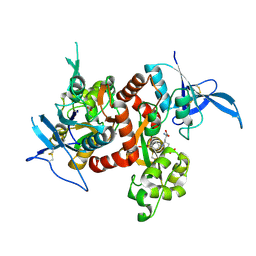 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-bromophenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-(4-bromophenyl)-1H-pyrazole-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2017-04-16 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TY3
 
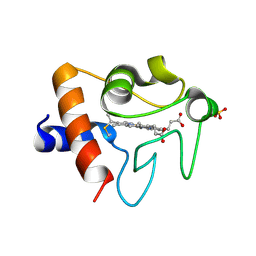 | | Crystal structure of K72A variant of Human Cytochrome c | | Descriptor: | Cytochrome c, HEME C, SULFATE ION | | Authors: | Mou, T.C, Nold, S.M, Lei, H, Sprang, S.R, Bowler, B.E. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Effect of a K72A Mutation on the Structure, Stability, Dynamics, and Peroxidase Activity of Human Cytochrome c.
Biochemistry, 56, 2017
|
|
1GG2
 
 | |
1EXT
 
 | |
1IK7
 
 | | Crystal Structure of the Uncomplexed Pelle Death Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PROBABLE SERINE/THREONINE-PROTEIN KINASE Pelle | | Authors: | Xiao, T, Gardner, K.H, Sprang, S.R. | | Deposit date: | 2001-05-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cosolvent-induced transformation
of a death domain tertiary structure
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1CJV
 
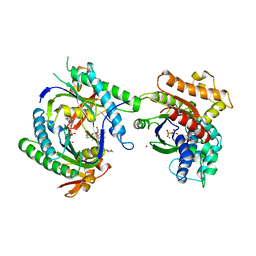 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH BETA-L-2',3'-DIDEOXYATP, MG, AND ZN | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1999-04-19 | | Release date: | 1999-08-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two-metal-Ion catalysis in adenylyl cyclase.
Science, 285, 1999
|
|
1CS4
 
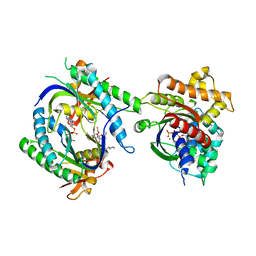 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH 2'-DEOXY-ADENOSINE 3'-MONOPHOSPHATE, PYROPHOSPHATE AND MG | | Descriptor: | 2'-DEOXY-ADENOSINE 3'-MONOPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Dessauer, C.A, Sunahara, R.K, Johnson, R.A, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1999-08-16 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for P-site inhibition of adenylyl cyclase.
Biochemistry, 39, 2000
|
|
1CJK
 
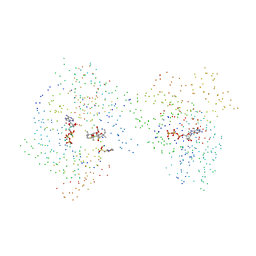 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH ADENOSINE 5'-(ALPHA THIO)-TRIPHOSPHATE (RP), MG, AND MN | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADENOSINE-5'-RP-ALPHA-THIO-TRIPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1999-04-16 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two-metal-Ion catalysis in adenylyl cyclase.
Science, 285, 1999
|
|
1CJT
 
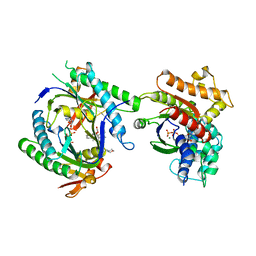 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH BETA-L-2',3'-DIDEOXYATP, MN, AND MG | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1999-04-16 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two-metal-Ion catalysis in adenylyl cyclase.
Science, 285, 1999
|
|
1CJU
 
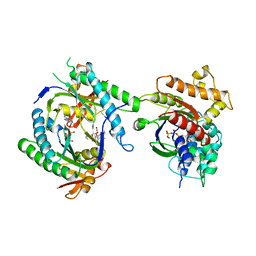 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH BETA-L-2',3'-DIDEOXYATP AND MG | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADENYLATE CYCLASE, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1999-04-16 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two-metal-Ion catalysis in adenylyl cyclase.
Science, 285, 1999
|
|
1CIP
 
 | |
1AS0
 
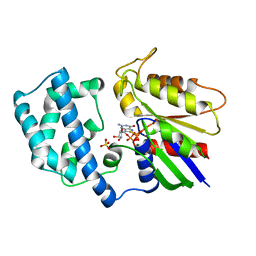 | | GTP-GAMMA-S BOUND G42V GIA1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GIA1, MAGNESIUM ION, ... | | Authors: | Raw, A.S, Coleman, D.E, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1997-08-11 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42 --> Val mutant of Gialpha1.
Biochemistry, 36, 1997
|
|
1AS3
 
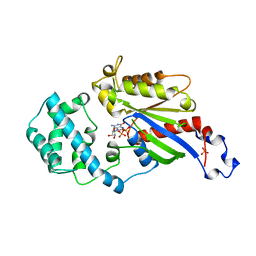 | | GDP BOUND G42V GIA1 | | Descriptor: | GIA1, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Raw, A.S, Coleman, D.E, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1997-08-11 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42 --> Val mutant of Gialpha1.
Biochemistry, 36, 1997
|
|
1AS2
 
 | | GDP+PI BOUND G42V GIA1 | | Descriptor: | GIA1, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Raw, A.S, Coleman, D.E, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1997-08-11 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42 --> Val mutant of Gialpha1.
Biochemistry, 36, 1997
|
|
1D2Z
 
 | | THREE-DIMENSIONAL STRUCTURE OF A COMPLEX BETWEEN THE DEATH DOMAINS OF PELLE AND TUBE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DEATH DOMAIN OF PELLE, DEATH DOMAIN OF TUBE | | Authors: | Xiao, T, Towb, P, Wasserman, S.A, Sprang, S.R. | | Deposit date: | 1999-09-28 | | Release date: | 1999-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a complex between the death domains of Pelle and Tube.
Cell(Cambridge,Mass.), 99, 1999
|
|
1AZT
 
 | | GS-ALPHA COMPLEXED WITH GTP-GAMMA-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GS-ALPHA, MAGNESIUM ION, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1997-11-20 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the adenylyl cyclase activator Gsalpha
Science, 278, 1997
|
|
1CUL
 
 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH 2',5'-DIDEOXY-ADENOSINE 3'-TRIPHOSPHATE AND MG | | Descriptor: | 2',5'-DIDEOXY-ADENOSINE 3'-MONOPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Dessauer, C.A, Sunahara, R.K, Johnson, R.A, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1999-08-20 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for P-site inhibition of adenylyl cyclase.
Biochemistry, 39, 2000
|
|
1AZS
 
 | |
1AGR
 
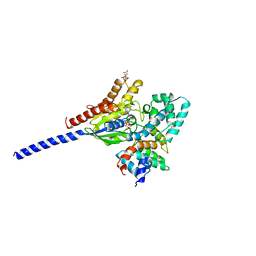 | | COMPLEX OF ALF4-ACTIVATED GI-ALPHA-1 WITH RGS4 | | Descriptor: | CITRIC ACID, GUANINE NUCLEOTIDE-BINDING PROTEIN G(I), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1997-03-25 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of RGS4 bound to AlF4--activated G(i alpha1): stabilization of the transition state for GTP hydrolysis.
Cell(Cambridge,Mass.), 89, 1997
|
|
1BH2
 
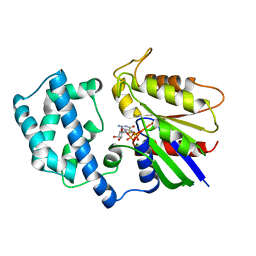 | | A326S MUTANT OF AN INHIBITORY ALPHA SUBUNIT | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANINE NUCLEOTIDE-BINDING PROTEIN, MAGNESIUM ION | | Authors: | Mixon, M.B, Posner, B.A, Wall, M.A, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1998-06-12 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The A326S mutant of Gialpha1 as an approximation of the receptor-bound state.
J.Biol.Chem., 273, 1998
|
|
1ANN
 
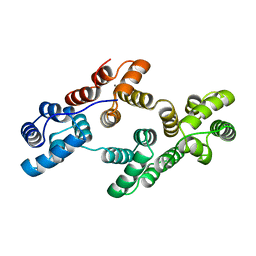 | | ANNEXIN IV | | Descriptor: | ANNEXIN IV, CALCIUM ION | | Authors: | Sutton, R.B, Sprang, S.R. | | Deposit date: | 1995-09-21 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three Dimensional Structure of Annexin IV
Annexins: Molecular Structure to Cellular Function, 1996
|
|
1A25
 
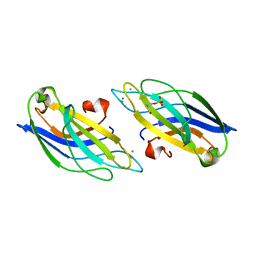 | | C2 DOMAIN FROM PROTEIN KINASE C (BETA) | | Descriptor: | CALCIUM ION, O-PHOSPHOETHANOLAMINE, PROTEIN KINASE C (BETA) | | Authors: | Sutton, R.B, Sprang, S.R. | | Deposit date: | 1998-01-16 | | Release date: | 1998-05-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the protein kinase Cbeta phospholipid-binding C2 domain complexed with Ca2+.
Structure, 6, 1998
|
|
1EDY
 
 | |
