4ET7
 
 | |
3OX4
 
 | | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 complexed with NAD cofactor | | Descriptor: | Alcohol dehydrogenase 2, FE (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Moon, J.H, Lee, H.J, Song, J.M, Park, S.Y, Park, M.Y, Park, H.M, Sun, J, Park, J.H, Kim, J.S. | | Deposit date: | 2010-09-21 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor
J.Mol.Biol., 407, 2011
|
|
3SXN
 
 | | Mycobacterium tuberculosis Eis protein initiates modulation of host immune responses by acetylation of DUSP16/MKP-7 | | Descriptor: | COENZYME A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J.S, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-07-15 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3OWO
 
 | | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD cofactor | | Descriptor: | Alcohol dehydrogenase 2, FE (II) ION | | Authors: | Moon, J.H, Lee, H.J, Song, J.M, Park, S.Y, Park, M.Y, Park, H.M, Sun, J, Park, J.H, Kim, J.S. | | Deposit date: | 2010-09-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor
J.Mol.Biol., 407, 2011
|
|
3FHA
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, GLYCEROL, ... | | Authors: | Yin, J, Li, L, Shaw, N, Li, Y, Song, J.K, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.C, Wang, L.X, Wang, P, Liu, Z.J. | | Deposit date: | 2008-12-09 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A.
Plos One, 4, 2009
|
|
1YYC
 
 | |
1Z3K
 
 | |
4RL5
 
 | |
2QCY
 
 | |
3OPE
 
 | | Structural Basis of Auto-inhibitory mechanism of Histone methyltransferase | | Descriptor: | Probable histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | An, S, Song, J. | | Deposit date: | 2010-08-31 | | Release date: | 2011-01-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism
J.Biol.Chem., 286, 2011
|
|
2FB7
 
 | |
3GL6
 
 | |
3EAJ
 
 | |
3EA9
 
 | |
3EA7
 
 | |
3EA8
 
 | |
4L67
 
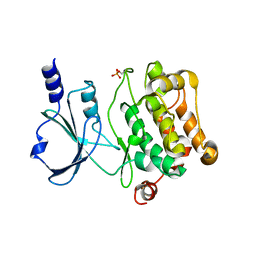 | | Crystal Structure of Catalytic Domain of PAK4 | | Descriptor: | Serine/threonine-protein kinase PAK 4 | | Authors: | Wang, W, Song, J. | | Deposit date: | 2013-06-12 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NMR binding and crystal structure reveal that intrinsically-unstructured regulatory domain auto-inhibits PAK4 by a mechanism different for that of PAK1
Biochem.Biophys.Res.Commun., 438, 2013
|
|
3NNH
 
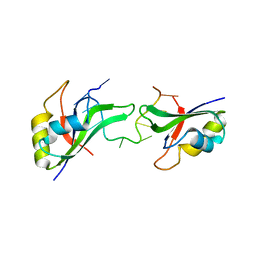 | | Crystal Structure of the CUGBP1 RRM1 with GUUGUUUUGUUU RNA | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7501 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
5CH6
 
 | |
3E91
 
 | |
8TDR
 
 | |
8TE3
 
 | |
8TE1
 
 | |
8TE4
 
 | |
3NNC
 
 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*UP*GP*UP*GP*UP*GP*UP*UP*GP*UP*GP*UP*G)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2005 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
