6LZG
 
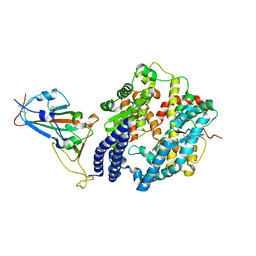 | | Structure of novel coronavirus spike receptor-binding domain complexed with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Wang, Q.H, Song, H, Qi, J.X. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2.
Cell, 181, 2020
|
|
7X0F
 
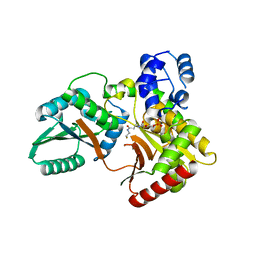 | |
7X0E
 
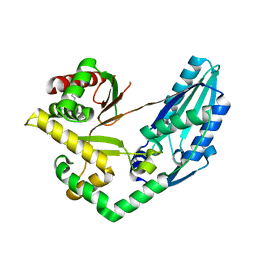 | | Structure of Pseudomonas NRPS protein, AmbB-TC in apo form | | Descriptor: | AMB antimetabolite synthase AmbB, N-methyl-N-[(2S,3R,4R,5R)-2,3,4,5,6-pentakis(oxidanyl)hexyl]nonanamide | | Authors: | ChuYuanKee, M, Bharath, S.R, Song, H. | | Deposit date: | 2022-02-22 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
7X17
 
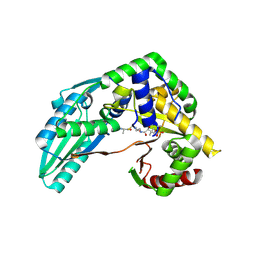 | | Structure of Pseudomonas NRPS protein, AmbB-TC bound to Ppant-L-Ala | | Descriptor: | AMB antimetabolite synthase AmbB, S-[2-[3-[[(2S)-3,3-dimethyl-2-oxidanyl-4-phosphonooxy-butanoyl]amino]propanoylamino]ethyl] (2R)-2-azanylpropanethioate | | Authors: | ChuYuanKee, M, Bharath, S.R, Song, H. | | Deposit date: | 2022-02-23 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
5XGR
 
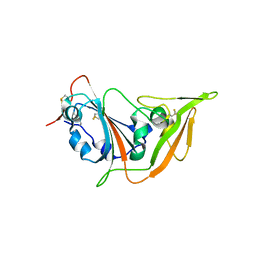 | | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Xue, H, Qi, J, Song, H, Qihui, W, Shi, Y, Gao, G.F. | | Deposit date: | 2017-04-16 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein
Virology, 507, 2017
|
|
7TRC
 
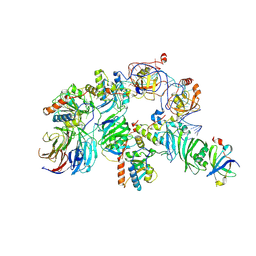 | | Human telomerase H/ACA RNP at 3.3 Angstrom | | Descriptor: | H/ACA ribonucleoprotein complex subunit 1, H/ACA ribonucleoprotein complex subunit 2, H/ACA ribonucleoprotein complex subunit 3, ... | | Authors: | Liu, B, He, Y, Wang, Y, Song, H, Zhou, Z.H, Feigon, J. | | Deposit date: | 2022-01-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of active human telomerase with telomere shelterin protein TPP1.
Nature, 604, 2022
|
|
7TRF
 
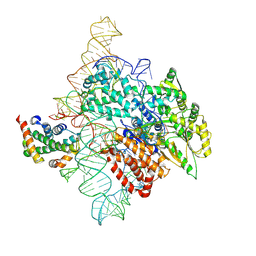 | | Human telomerase catalytic core RNP with H2A/H2B | | Descriptor: | Histone H2A, Histone H2B type 1-C/E/F/G/I, Telomerase RNA, ... | | Authors: | Liu, B, He, Y, Wang, Y, Song, H, Zhou, Z.H, Feigon, J. | | Deposit date: | 2022-01-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of active human telomerase with telomere shelterin protein TPP1.
Nature, 604, 2022
|
|
7TRD
 
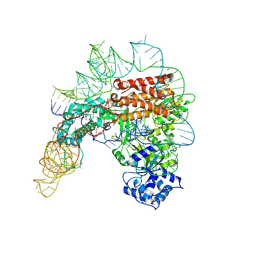 | | Human telomerase catalytic core structure at 3.3 Angstrom | | Descriptor: | Telomerase RNA, partial sequence, Telomerase reverse transcriptase, ... | | Authors: | Liu, B, He, Y, Wang, Y, Song, H, Zhou, Z.H, Feigon, J. | | Deposit date: | 2022-01-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of active human telomerase with telomere shelterin protein TPP1.
Nature, 604, 2022
|
|
7TRE
 
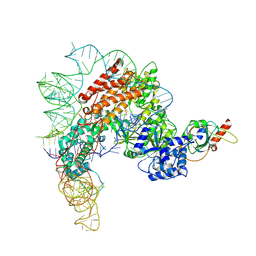 | | Human telomerase catalytic core with shelterin protein TPP1 | | Descriptor: | Adrenocortical dysplasia protein homolog, Telomerase RNA, partial sequence, ... | | Authors: | Liu, B, He, Y, Wang, Y, Song, H, Zhou, Z.H, Feigon, J. | | Deposit date: | 2022-01-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of active human telomerase with telomere shelterin protein TPP1.
Nature, 604, 2022
|
|
7BZT
 
 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZN
 
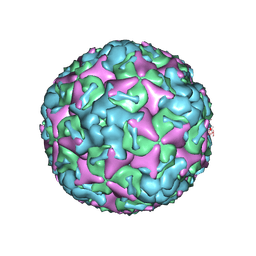 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZO
 
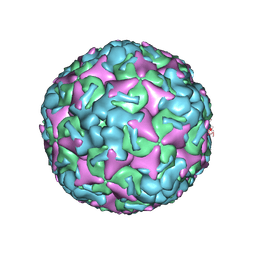 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZU
 
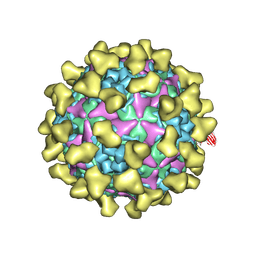 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BV7
 
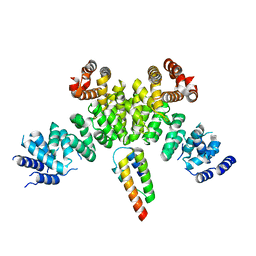 | | INTS3 complexed with INTS6 | | Descriptor: | Integrator complex subunit 3, Integrator complex subunit 6 | | Authors: | Jia, Y, Bharath, S.R, Song, H. | | Deposit date: | 2020-04-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the INTS3/INTS6 complex reveals the functional importance of INTS3 dimerization in DSB repair.
Cell Discov, 7, 2021
|
|
7C4Z
 
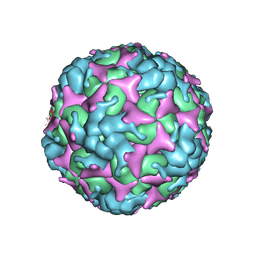 | | Cryo-EM structure of empty Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4Y
 
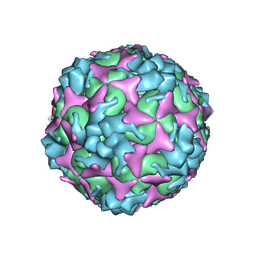 | | Cryo-EM structure of empty Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CDZ
 
 | |
7C4T
 
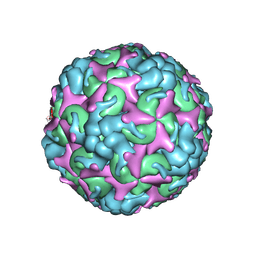 | | Cryo-EM structure of A particle Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CE0
 
 | |
7C4W
 
 | | Cryo-EM structure of A particle Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5XSQ
 
 | | Crystal Structure of the Marburg Virus Nucleoprotein Core Domain Chaperoned by a VP35 Peptide | | Descriptor: | Nucleoprotein, Peptide from Polymerase cofactor VP35 | | Authors: | Zhu, T, Song, H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-06-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Marburg Virus Nucleoprotein Core Domain Chaperoned by a VP35 Peptide Reveals a Conserved Drug Target for Filovirus
J. Virol., 91, 2017
|
|
7CZF
 
 | | Crystal structure of Kaposi Sarcoma associated herpesvirus (KSHV ) gHgL in complex with the ligand binding domian (LBD) of EphA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Su, C, Wu, L.L, Song, H, Chai, Y, Qi, J.X, Yan, J.H, Gao, G.F. | | Deposit date: | 2020-09-08 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis of EphA2 recognition by gHgL from gammaherpesviruses.
Nat Commun, 11, 2020
|
|
7CZE
 
 | | Crystal structure of Epstein-Barr virus (EBV) gHgL and in complex with the ligand binding domian (LBD) of EphA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Su, C, Wu, L.L, Song, H, Chai, Y, Qi, J.X, Yan, J.H, Gao, G.F. | | Deposit date: | 2020-09-08 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of EphA2 recognition by gHgL from gammaherpesviruses.
Nat Commun, 11, 2020
|
|
5GHV
 
 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-({1-[2-({3,5-dimethyl-4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]-4-methyl-1H-pyrrol-3-yl}methyl)azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
5YGH
 
 | | Crystal Structure of the Capsid Protein from Zika Virus | | Descriptor: | Capsid protein | | Authors: | Shang, Z, Song, H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-09-23 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Crystal Structure of the Capsid Protein from Zika Virus.
J. Mol. Biol., 430, 2018
|
|
