5RLN
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z364328788 | | 分子名称: | 3-(acetylamino)-4-fluorobenzoic acid, Helicase, PHOSPHATE ION, ... | | 著者 | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-09-16 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.147 Å) | | 主引用文献 | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RM5
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z373768900 | | 分子名称: | Helicase, N-(1-ethyl-1H-pyrazol-4-yl)cyclobutanecarboxamide, PHOSPHATE ION, ... | | 著者 | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-09-16 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RMI
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z53860899 | | 分子名称: | Helicase, PHOSPHATE ION, ZINC ION, ... | | 著者 | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-09-16 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.116 Å) | | 主引用文献 | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLL
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z425387594 | | 分子名称: | 1-(2-ethoxyphenyl)piperazine, Helicase, PHOSPHATE ION, ... | | 著者 | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-09-16 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLV
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z2467208649 | | 分子名称: | Helicase, N-(propan-2-yl)-1H-benzimidazol-2-amine, PHOSPHATE ION, ... | | 著者 | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-09-16 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RM8
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1614545742 | | 分子名称: | Helicase, PHOSPHATE ION, ZINC ION, ... | | 著者 | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-09-16 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.143 Å) | | 主引用文献 | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLF
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z235341991 | | 分子名称: | Helicase, N-(2-methoxy-5-methylphenyl)glycinamide, PHOSPHATE ION, ... | | 著者 | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-09-16 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.235 Å) | | 主引用文献 | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLU
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z744754722 | | 分子名称: | 2-(thiophen-2-yl)-1H-imidazole, Helicase, PHOSPHATE ION, ... | | 著者 | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-09-16 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.347 Å) | | 主引用文献 | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RME
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z26333434 | | 分子名称: | 4-(benzimidazol-1-ylmethyl)benzenecarbonitrile, Helicase, PHOSPHATE ION, ... | | 著者 | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-09-16 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
4D08
 
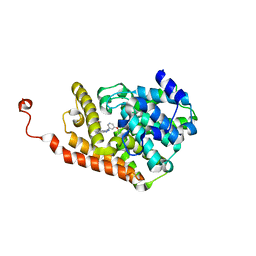 | | PDE2a catalytic domain in complex with a brain penetrant inhibitor | | 分子名称: | 1-(5-butoxypyridin-3-yl)-4-methyl-8-(morpholin-4-ylmethyl)[1,2,4]triazolo[4,3-a]quinoxaline, CGMP-DEPENDENT 3', 5'-CYCLIC PHOSPHODIESTERASE, ... | | 著者 | Buijnsters, P, Andres, J.I, DeAngelis, M, Langlois, X, Rombouts, F, Sanderson, W, Tresadern, G, Trabanco, A, VanHoof, G, VanRoosbroeck, Y. | | 登録日 | 2014-04-24 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Based Design of a Potent, Selective, and Brain Penetrating Pde2 Inhibitor with Demonstrated Target Engagement.
Acs Med.Chem.Lett., 5, 2014
|
|
4D09
 
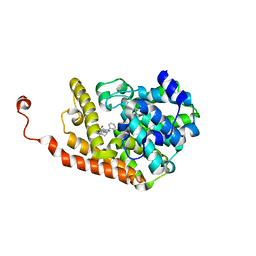 | | PDE2a catalytic domain in complex with a brain penetrant inhibitor | | 分子名称: | CGMP-DEPENDENT 3', 5'-CYCLIC PHOSPHODIESTERASE, MAGNESIUM ION, ... | | 著者 | Buijnsters, P, Andres, J.I, DeAngelis, M, Langlois, X, Rombouts, F, Sanderson, W, Tresadern, G, Trabanco, A, VanHoof, G, VanRoosbroeck, Y. | | 登録日 | 2014-04-24 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-Based Design of a Potent, Selective, and Brain Penetrating Pde2 Inhibitor with Demonstrated Target Engagement.
Acs Med.Chem.Lett., 5, 2014
|
|
2LRJ
 
 | |
2M1Z
 
 | |
5TVL
 
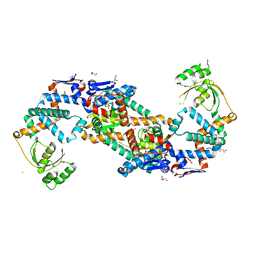 | | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A | | 分子名称: | CHLORIDE ION, Foldase protein PrsA, GLYCEROL, ... | | 著者 | Borek, D, Yim, V, Kudritska, M, Wawrzak, Z, Stogios, P.J, Otwinowski, Z, Savchenko, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-11-09 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A
To Be Published
|
|
2L3M
 
 | | Solution structure of the putative copper-ion-binding protein from Bacillus anthracis str. Ames | | 分子名称: | Copper-ion-binding protein | | 著者 | Zhang, Y, Winsor, J, Dubrovska, I, Anderson, W, Radhakrishnan, I, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-09-16 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | To be published
To be Published
|
|
7ETW
 
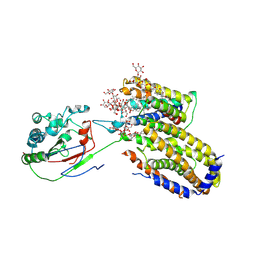 | | Cryo-EM structure of Scap/Insig complex in the present of digitonin. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Digitonin, Insulin-induced gene 2 protein, ... | | 著者 | Yan, R, Cao, P, Song, W, Li, Y, Wang, T, Qian, H, Yan, C, Yan, N. | | 登録日 | 2021-05-14 | | 公開日 | 2021-06-23 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis for sterol sensing by Scap and Insig
Cell Rep, 35, 2021
|
|
7F18
 
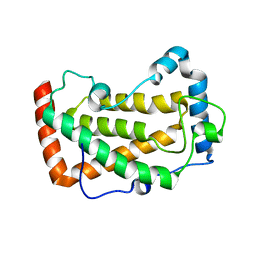 | | Crystal Structure of a mutant of acid phosphatase from Pseudomonas aeruginosa (Q57H/W58P/D135R) | | 分子名称: | Acid phosphatase | | 著者 | Xu, X, Hou, X.D, Song, W, Yin, D.J, Rao, Y.J, Liu, L.M. | | 登録日 | 2021-06-08 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Local Electric Field Modulated Reactivity of Pseudomonas aeruginosa Acid Phosphatase for Enhancing Phosphorylation of l-Ascorbic Acid
Acs Catalysis, 11, 2021
|
|
7F17
 
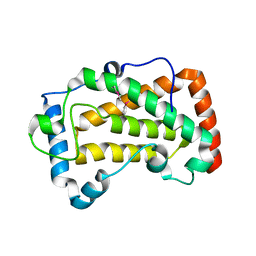 | | Crystal Structure of acid phosphatase | | 分子名称: | Acid phosphatase | | 著者 | Xu, X, Hou, X.D, Song, W, Rao, Y.J, Liu, L.M, Wu, J. | | 登録日 | 2021-06-08 | | 公開日 | 2021-10-27 | | 最終更新日 | 2022-05-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Local Electric Field Modulated Reactivity of Pseudomonas aeruginosa Acid Phosphatase for Enhancing Phosphorylation of l-Ascorbic Acid
Acs Catalysis, 11, 2021
|
|
1UFH
 
 | | Structure of putative acetyltransferase, YYCN protein of Bacillus subtilis | | 分子名称: | YYCN protein | | 著者 | Taneja, B, Maar, S, Shuvalova, L, Collart, F.R, Anderson, W, Mondragon, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-05-29 | | 公開日 | 2003-06-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the bacillus subtilis YYCN protein: a putative N-acetyltransferase
Proteins, 53, 2003
|
|
1MS6
 
 | | Dipeptide Nitrile Inhibitor Bound to Cathepsin S. | | 分子名称: | Cathepsin S, MORPHOLINE-4-CARBOXYLIC ACID [1S-(2-BENZYLOXY-1R-CYANO-ETHYLCARBAMOYL)-3-METHYL-BUTYL]AMIDE | | 著者 | Ward, Y.D, Thomson, D.S, Frye, L.L, Cywin, C.L, Morwick, T, Emmanuel, M.J, Zindell, R, McNeil, D, Bekkali, Y, Giradot, M, Hrapchak, M, DeTuri, M, Crane, K, White, D, Pav, S, Wang, Y, Hao, M.H, Grygon, C.A, Labadia, M.E, Freeman, D.M, Davidson, W, Hopkins, J.L, Brown, M.L, Spero, D.M. | | 登録日 | 2002-09-19 | | 公開日 | 2003-04-22 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Design and synthesis of dipeptide nitriles as reversible and potent Cathepsin S inhibitors
J.Med.Chem., 45, 2002
|
|
4BBX
 
 | | Discovery of a potent, selective and orally active PDE10A inhibitor for the treatment of schizophrenia | | 分子名称: | 4-[3-[1-[(2S)-2-methoxypropyl]pyrazol-4-yl]-2-methyl-imidazo[1,2-a]pyrazin-8-yl]morpholine, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | 著者 | Bartolome-Nebreda, J.M, Conde-Ceide, S, Delgado, F, Martin, M.L, Martinez-Viturro, C.M, Pastor, J, Tong, H.M, Iturrino, L, Macdonald, G.J, Sanderson, W, Megens, A, Langlois, X, Somers, M, Vanhoof, G. | | 登録日 | 2012-09-28 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of a Potent, Selective and Orally Active Pde10A Inhibitor for the Potential Treatment of Schizophrenia.
J.Med.Chem., 57, 2014
|
|
2LWG
 
 | |
2LWH
 
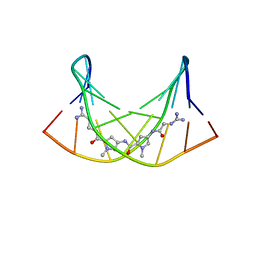 | | NMR Structure of the Self-Complementary 10 mer DNA Duplex 5'-GGATATATCC-3' in Complex with Netropsin | | 分子名称: | DNA (5'-D(*GP*GP*AP*TP*AP*TP*AP*TP*CP*C)-3'), NETROPSIN | | 著者 | Rettig, M, Germann, M.W, Wilson, W, Wang, S. | | 登録日 | 2012-07-31 | | 公開日 | 2013-01-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular basis for sequence-dependent induced DNA bending.
Chembiochem, 14, 2013
|
|
2J8U
 
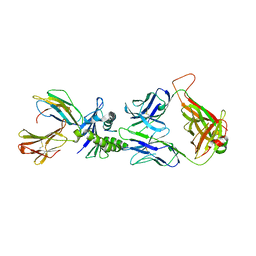 | | Large CDR3a loop alteration as a function of MHC mutation. | | 分子名称: | AHIII TCR ALPHA CHAIN, AHIII TCR BETA CHAIN, Beta-2-microglobulin, ... | | 著者 | Miller, P.J, Benhar, Y.P, Biddison, W, Collins, E.J. | | 登録日 | 2006-10-27 | | 公開日 | 2007-10-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Single MHC mutation eliminates enthalpy associated with T cell receptor binding.
J. Mol. Biol., 373, 2007
|
|
2JCC
 
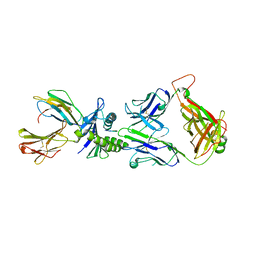 | | AH3 recognition of mutant HLA-A2 W167A | | 分子名称: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-2 ALPHA CHAIN, ... | | 著者 | Miller, P, Benhar, Y.P, Biddison, W, Collins, E.J. | | 登録日 | 2006-12-21 | | 公開日 | 2007-10-09 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Single Mhc Mutation Eliminates Enthalpy Associated with T Cell Receptor Binding.
J.Mol.Biol., 373, 2007
|
|
