1MDX
 
 | | Crystal structure of ArnB transferase with pyridoxal 5' phosphate | | Descriptor: | 2-OXOGLUTARIC ACID, ArnB aminotransferase, GLYCEROL, ... | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T.A, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K.S, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
3RNG
 
 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase T201S/W167E Double Mutant | | Descriptor: | FE (III) ION, HYDROXIDE ION, Toluene o-xylene monooxygenase component | | Authors: | Gucinski, G, Song, W.J, Lippard, S.J, Sazinsky, M.H. | | Deposit date: | 2011-04-22 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Tracking a defined route for O2 migration in a dioxygen-activating diiron enzyme.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2LRJ
 
 | |
9FGO
 
 | | Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site | | Descriptor: | CHLORIDE ION, Polyprotein, ZINC ION | | Authors: | Ni, X, Koekemoer, L, Williams, E.P, Wang, S, Wright, N.D, Godoy, A.S, Aschenbrenner, J.C, Balcomb, B.H, Lithgo, R.M, Marples, P.G, Fairhead, M, Thompson, W, Kirkegaard, K, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-05-24 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site
To Be Published
|
|
1IRK
 
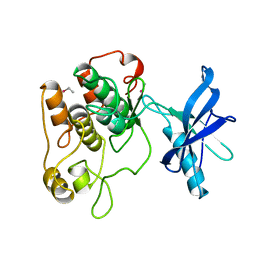 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF THE HUMAN INSULIN RECEPTOR | | Descriptor: | ETHYL MERCURY ION, INSULIN RECEPTOR TYROSINE KINASE DOMAIN | | Authors: | Hubbard, S.R, Wei, L, Ellis, L, Hendrickson, W.A. | | Deposit date: | 1995-01-02 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the tyrosine kinase domain of the human insulin receptor.
Nature, 372, 1994
|
|
1INN
 
 | | CRYSTAL STRUCTURE OF D. RADIODURANS LUXS, P21 | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
1J6V
 
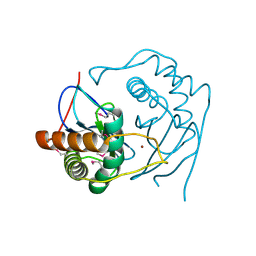 | | CRYSTAL STRUCTURE OF D. RADIODURANS LUXS, C2 | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
1J6W
 
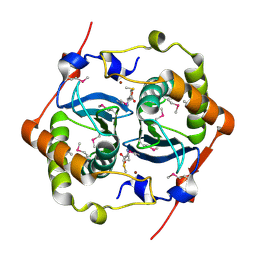 | | CRYSTAL STRUCTURE OF HAEMOPHILUS INFLUENZAE LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
7K7R
 
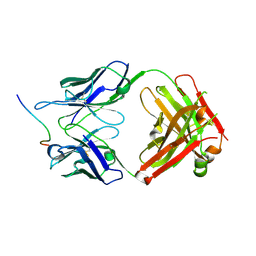 | | EBNA1 peptide AA386-405 with Fab MS39p2w174 | | Descriptor: | CHLORIDE ION, EBNA1 peptide AA386-405, Fab HC MS39p2w174, ... | | Authors: | Lanz, T.V, Robinson, W.H, Jude, K.M. | | Deposit date: | 2020-09-23 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Clonally expanded B cells in multiple sclerosis bind EBV EBNA1 and GlialCAM.
Nature, 603, 2022
|
|
2M1Z
 
 | |
1BDO
 
 | |
2NSB
 
 | |
2NSA
 
 | |
7JO8
 
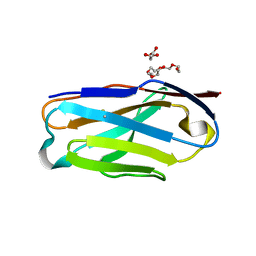 | | Crystal structure of a Chimeric Antigen Receptor (CAR) scFv domain rearrangement forming a VL-VL dimer | | Descriptor: | 47G4-CD828Z, MALONATE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Cheung, J, Hendrickson, W.A, Kochenderfer, J.N, Youkharibache, P. | | Deposit date: | 2020-08-06 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Crystal Structure of a Chimeric Antigen Receptor (CAR) scFv Domain Rearrangement Forming a VL-VL Dimer
Crystals, 13, 2023
|
|
2NSC
 
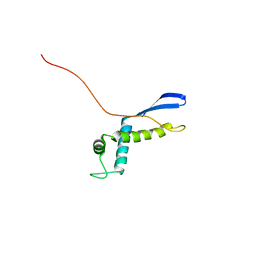 | |
2OKQ
 
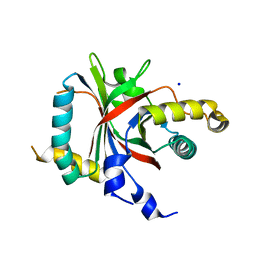 | | Crystal structure of unknown conserved ybaA protein from Shigella flexneri | | Descriptor: | Hypothetical protein ybaA, SODIUM ION | | Authors: | Minasov, G, Vorontsov, I.I, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-17 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of unknown conserved ybaA protein from Shigella flexneri
TO BE PUBLISHED
|
|
2OL5
 
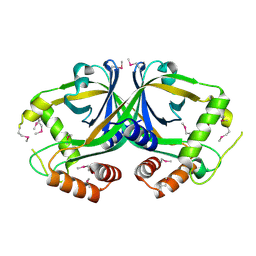 | | Crystal Structure of a protease synthase and sporulation negative regulatory protein PAI 2 from Bacillus stearothermophilus | | Descriptor: | PAI 2 protein | | Authors: | Brunzelle, J.S, Cuff, M.E, Minasov, G, Li, H, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-18 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the novel PaiB transcriptional regulator from Geobacillus stearothermophilus.
Proteins, 79, 2011
|
|
1AXM
 
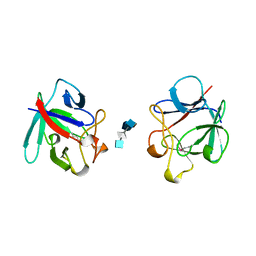 | | HEPARIN-LINKED BIOLOGICALLY-ACTIVE DIMER OF FIBROBLAST GROWTH FACTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | DiGabriele, A.D, Lax, I, Chen, D.I, Svahn, C.M, Jaye, M, Schlessinger, J, Hendrickson, W.A. | | Deposit date: | 1997-10-16 | | Release date: | 1998-04-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a heparin-linked biologically active dimer of fibroblast growth factor.
Nature, 393, 1998
|
|
1L7C
 
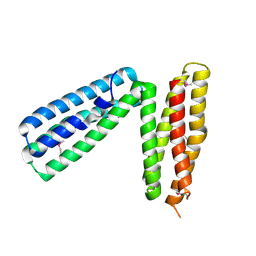 | | alpha-catenin fragment, residues 385-651 | | Descriptor: | Alpha E-catenin | | Authors: | Pokutta, S, Drees, F, Takai, Y, Nelson, W.J, Weis, W.I. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and structural definition of the l-afadin- and actin-binding sites of alpha-catenin.
J.Biol.Chem., 277, 2002
|
|
2PR1
 
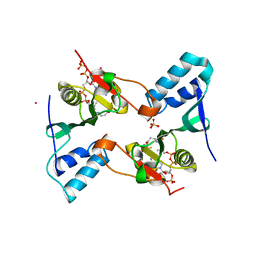 | | Crystal structure of the Bacillus subtilis N-acetyltransferase YlbP protein in complex with Coenzyme-A | | Descriptor: | COBALT (II) ION, COENZYME A, SULFATE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Vorontsov, I.I, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-03 | | Release date: | 2007-05-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the Bacillus subtilis N-acetyltransferase YlbP protein in complex with Coenzyme-A.
To be Published
|
|
1CNT
 
 | | CILIARY NEUROTROPHIC FACTOR | | Descriptor: | CILIARY NEUROTROPHIC FACTOR, SULFATE ION, YTTERBIUM (III) ION | | Authors: | Mcdonald, N.Q, Panayotatos, N, Hendrickson, W.A. | | Deposit date: | 1996-06-06 | | Release date: | 1997-03-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of dimeric human ciliary neurotrophic factor determined by MAD phasing.
EMBO J., 14, 1995
|
|
1BUN
 
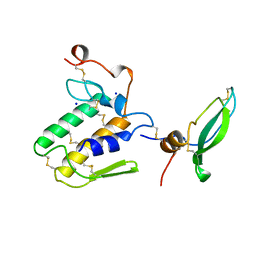 | | STRUCTURE OF BETA2-BUNGAROTOXIN: POTASSIUM CHANNEL BINDING BY KUNITZ MODULES AND TARGETED PHOSPHOLIPASE ACTION | | Descriptor: | BETA2-BUNGAROTOXIN, SODIUM ION | | Authors: | Kwong, P.D, Mcdonald, N.Q, Sigler, P.B, Hendrickson, W.A. | | Deposit date: | 1995-10-15 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of beta 2-bungarotoxin: potassium channel binding by Kunitz modules and targeted phospholipase action.
Structure, 3, 1995
|
|
1J6X
 
 | | CRYSTAL STRUCTURE OF HELICOBACTER PYLORI LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
2Q6T
 
 | |
1ITH
 
 | | STRUCTURE DETERMINATION AND REFINEMENT OF HOMOTETRAMERIC HEMOGLOBIN FROM URECHIS CAUPO AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | CYANIDE ION, HEMOGLOBIN (CYANO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hackert, M, Kolatkar, P, Ernst, S.R, Ogata, C.M, Hendrickson, W.A, Merritt, E.A, Phizackerley, R.P. | | Deposit date: | 1991-12-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination and refinement of homotetrameric hemoglobin from Urechis caupo at 2.5 A resolution.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
