3GSD
 
 | | 2.05 Angstrom structure of a divalent-cation tolerance protein (CutA) from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Structure of a Divalent-cation Tolerance Protein (CutA) from Yersinia pestis
TO BE PUBLISHED
|
|
4XDN
 
 | | Crystal structure of Scc4 in complex with Scc2n | | Descriptor: | MAU2 chromatid cohesion factor homolog, SULFATE ION, Sister chromatid cohesion protein 2 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2014-12-19 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural evidence for Scc4-dependent localization of cohesin loading.
Elife, 4, 2015
|
|
2W81
 
 | | Structure of a complex between Neisseria meningitidis factor H binding protein and CCPs 6-7 of human complement factor H | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Schneider, M.C, Prosser, B.E, Caesar, J.J.E, Kugelberg, E, Li, S, Zhang, Q, Quoraishi, S, Lovett, J.E, Deane, J.E, Sim, R.B, Roversi, P, Johnson, S, Tang, C.M, Lea, S.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Neisseria Meningitidis Recruits Factor H Using Protein Mimicry of Host Carbohydrates.
Nature, 458, 2009
|
|
3HID
 
 | | Crystal structure of adenylosuccinate synthetase from Yersinia pestis CO92 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Adenylosuccinate synthetase | | Authors: | Zhang, R, Zhou, M, Peterson, S, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-19 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the adenylosuccinate synthetase from Yersinia pestis CO92
To be Published
|
|
5NJG
 
 | | Structure of an ABC transporter: part of the structure that could be built de novo | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3-Fab heavy chain, 5D3-Fab light chain, ... | | Authors: | Taylor, N.M.I, Manolaridis, I, Jackson, S.M, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structure of the human multidrug transporter ABCG2.
Nature, 546, 2017
|
|
5KS5
 
 | | Structure of the C-terminal Helical Repeat Domain of Elongation Factor 2 Kinase | | Descriptor: | Eukaryotic elongation factor 2 kinase | | Authors: | Piserchio, A, Will, N, Snyder, I, Ferguson, S.B, Giles, D.H, Dalby, K.N, Ghose, R. | | Deposit date: | 2016-07-07 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-Terminal Helical Repeat Domain of Eukaryotic Elongation Factor 2 Kinase.
Biochemistry, 55, 2016
|
|
7TKV
 
 | | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Kim, Y, Crawford, M, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-17 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus
To Be Published
|
|
5J29
 
 | |
5J2G
 
 | |
5J0P
 
 | |
5J0S
 
 | |
5J2E
 
 | |
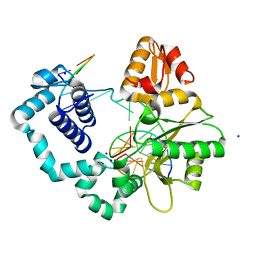
5J0U
 
 | |
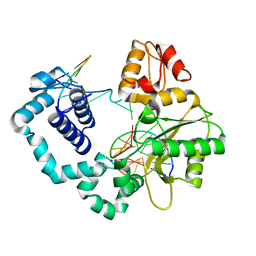
5J0Q
 
 | |
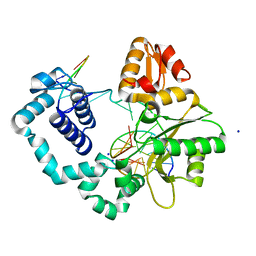
5J0T
 
 | |
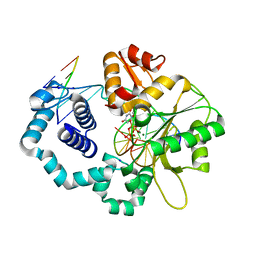
5J2I
 
 | |
2XRC
 
 | | Human complement factor I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HUMAN COMPLEMENT FACTOR I | | Authors: | Roversi, P, Johnson, S, Lea, S.M. | | Deposit date: | 2010-09-13 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural Basis for Complement Factor I Control and its Disease-Associated Sequence Polymorphisms.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1H1B
 
 | | Crystal structure of human neutrophil elastase complexed with an inhibitor (GW475151) | | Descriptor: | (2S)-3-METHYL-2-((2R,3S)-3-[(METHYLSULFONYL)AMINO]-1-{[2-(PYRROLIDIN-1-YLMETHYL)-1,3-OXAZOL-4-YL]CARBONYL}PYRROLIDIN-2-YL)BUTANOIC ACID, LEUKOCYTE ELASTASE, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Macdonald, S.J.F, Dowle, M.D, Harrison, L.A, Clarke, G.D.E, Inglis, G.G.A, Johnson, M.R, Smith, R.A, Amour, A, Fleetwood, G, Humphreys, D.C, Molloy, C.R, Dixon, M, Godward, R.E, Wonacott, A.J, Singh, O.M.P, Hodgson, S.T, Hardy, G.W. | | Deposit date: | 2002-07-05 | | Release date: | 2002-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Further Pyrrolidine Trans-Lactams as Inhibitors of Human Neutrophil Elastase (Hne) with Potential as Development Candidates and the Crystal Structure of Hne Complexed with an Inhibitor (Gw475151)
J.Med.Chem., 45, 2002
|
|
2XRB
 
 | | Structure of the N-terminal four domains of the complement regulator Rat Crry | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT REGULATORY PROTEIN CRRY, SULFATE ION | | Authors: | Leath, K.J, Roversi, P, Johnson, S, Morgan, B.P, Lea, S.M. | | Deposit date: | 2010-09-13 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the Rat Complement Regulator Crry.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
6X3L
 
 | | Sortilin-Progranulin Interaction With Compound 2 | | Descriptor: | 1-benzyl-3-tert-butyl-1H-pyrazole-5-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Parthasarathy, G, Soisson, S.M. | | Deposit date: | 2020-05-21 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of potent inhibitors of the sortilin-progranulin interaction.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6X48
 
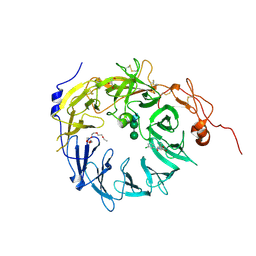 | | Sortilin-Progranulin Interaction With Compound 17 | | Descriptor: | GLYCEROL, N-(3,5-dichlorobenzene-1-carbonyl)-5,5-dimethyl-L-norleucine, Sortilin, ... | | Authors: | Parthasarathy, G, Soisson, S.M. | | Deposit date: | 2020-05-22 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of potent inhibitors of the sortilin-progranulin interaction.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5A2E
 
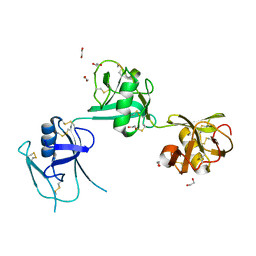 | | Extracellular SRCR domains of human CD6 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, T-CELL DIFFERENTIATION ANTIGEN CD6 | | Authors: | Chappell, P.E, Johnson, S, Lea, S.M, Brown, M.H. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of Cd6 and its Ligand Cd166 Give Insight Into Their Interaction.
Structure, 23, 2015
|
|
5J2A
 
 | |
5J2F
 
 | |
5AL7
 
 | |
