2R7V
 
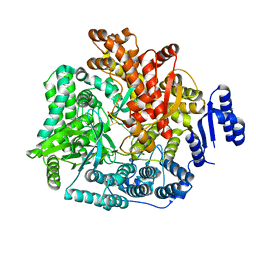 | | Crystal Structure of Rotavirus SA11 VP1/RNA (GGCUUU) Complex | | Descriptor: | RNA (5'-R(*G*GP*CP*UP*UP*U)-3'), RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
2BEL
 
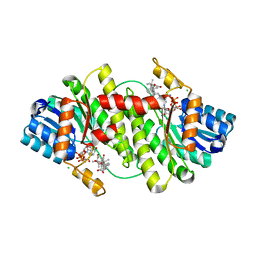 | | Structure of human 11-beta-hydroxysteroid dehydrogenase in complex with NADP and carbenoxolone | | Descriptor: | CARBENOXOLONE, CHLORIDE ION, CORTICOSTEROID 11-BETA-DEHYDROGENASE ISOZYME 1, ... | | Authors: | Kavanagh, K, Wu, X, Svensson, S, Elleby, B, von Delft, F, Debreczeni, J.E, Sharma, S, Bray, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Abrahmsen, L, Oppermann, U. | | Deposit date: | 2004-11-25 | | Release date: | 2004-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The High Resolution Structures of Human, Murine and Guinea Pig 11-Beta-Hydroxysteroid Dehydrogenase Type 1 Reveal Critical Differences in Active Site Architecture
To be Published
|
|
1JE8
 
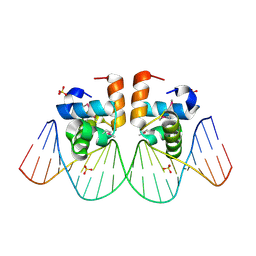 | | Two-Component response regulator NarL/DNA Complex: DNA Bending Found in a High Affinity Site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*CP*CP*AP*TP*TP*AP*AP*TP*GP*GP*GP*TP*AP*CP*G)-3', Nitrate/Nitrite Response Regulator Protein NARL, SULFATE ION | | Authors: | Maris, A.E, Sawaya, M.R, Kaczor-Grzeskowiak, M, Jarvis, M.R, Bearson, S.M.D, Kopka, M.L, Schroder, I, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 2001-06-15 | | Release date: | 2002-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Dimerization allows DNA target site recognition by the NarL response regulator.
Nat.Struct.Biol., 9, 2002
|
|
1XSX
 
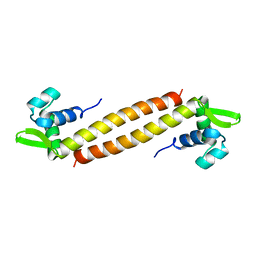 | | NMR Structure of Sso10a, a Hyperthermophile DNA-binding Protein with an Extended Anti-parallel Coiled Coil | | Descriptor: | Sso10a | | Authors: | Kahsai, M.A, Vogler, B, Clark, A.T, Edmondson, S.P, Shriver, J.W. | | Deposit date: | 2004-10-20 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Stability, and Flexibility of Sso10a: A Hyperthermophile Coiled-Coil DNA-Binding Protein(,).
Biochemistry, 44, 2005
|
|
2HFN
 
 | | Crystal Structures of the Synechocystis Photoreceptor Slr1694 Reveal Distinct Structural States Related to Signaling | | Descriptor: | FLAVIN MONONUCLEOTIDE, Synechocystis Photoreceptor (Slr1694) | | Authors: | Yuan, H, Anderson, S, Masuda, S, Dragnea, V, Moffat, K, Bauer, C.E. | | Deposit date: | 2006-06-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling.
Biochemistry, 45, 2006
|
|
2VML
 
 | |
1IGT
 
 | | STRUCTURE OF IMMUNOGLOBULIN | | Descriptor: | IGG2A INTACT ANTIBODY - MAB231, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Harris, L.J, Larson, S.B, Hasel, K.W, McPherson, A. | | Deposit date: | 1996-10-25 | | Release date: | 1997-07-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Refined structure of an intact IgG2a monoclonal antibody.
Biochemistry, 36, 1997
|
|
2HHF
 
 | |
3RIJ
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | GLYCEROL, SC_2cx5 | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-13 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
1SLQ
 
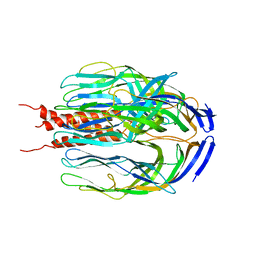 | | Crystal structure of the trimeric state of the rhesus rotavirus VP4 membrane interaction domain, VP5CT | | Descriptor: | VP4 | | Authors: | Dormitzer, P.R, Nason, E.B, Prasad, B.V.V, Harrison, S.C. | | Deposit date: | 2004-03-06 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural rearrangements in the membrane penetration protein of a non-enveloped virus.
Nature, 430, 2004
|
|
1JS9
 
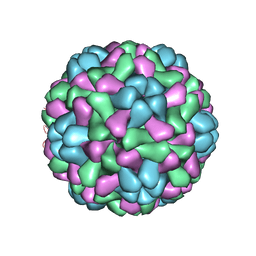 | | Brome Mosaic Virus | | Descriptor: | Coat protein, HEXAETHYLENE GLYCOL, MAGNESIUM ION | | Authors: | Lucas, R.W, Larson, S.B, McPherson, A. | | Deposit date: | 2001-08-16 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The crystallographic structure of brome mosaic virus.
J.Mol.Biol., 317, 2002
|
|
4HZA
 
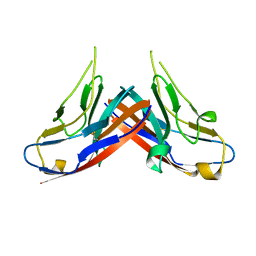 | |
1JUF
 
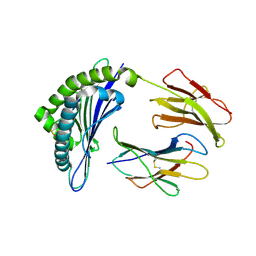 | | Structure of Minor Histocompatibility Antigen peptide, H13b, complexed to H2-Db | | Descriptor: | Beta-2-microglobulin, H13b peptide, H2-Db major histocompatibility antigen | | Authors: | Ostrov, D.A, Roden, M.M, Shi, W, Palmieri, E, Christianson, G.J, Mendoza, L, Villaflor, G, Tilley, D, Shastri, N, Grey, H, Almo, S.C, Roopenian, D.C, Nathenson, S.G. | | Deposit date: | 2001-08-24 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How H13 histocompatibility peptides differing by a single methyl group and lacking conventional MHC binding anchor motifs determine self-nonself discrimination.
J.Immunol., 168, 2002
|
|
2HFO
 
 | | Crystal Structures of the Synechocystis Photoreceptor Slr1694 Reveal Distinct Structural States Related to Signaling | | Descriptor: | Activator of photopigment and puc expression, FLAVIN MONONUCLEOTIDE | | Authors: | Yuan, H, Anderson, S, Masuda, S, Dragnea, V, Moffat, K, Bauer, C.E. | | Deposit date: | 2006-06-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling.
Biochemistry, 45, 2006
|
|
2HGW
 
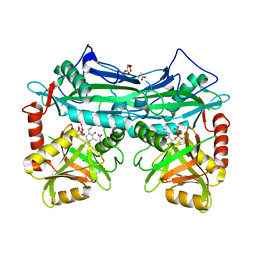 | |
1K42
 
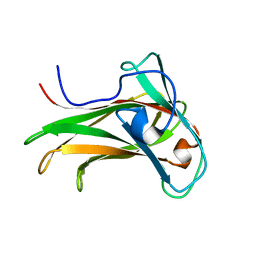 | | The Solution Structure of the CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase. | | Descriptor: | Xylanase | | Authors: | Simpson, P.J, Jamieson, S.J, Abou-Hachem, M, Nordberg-Karlsson, E, Gilbert, H.J, Holst, O, Williamson, M.P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase.
Biochemistry, 41, 2002
|
|
1KE5
 
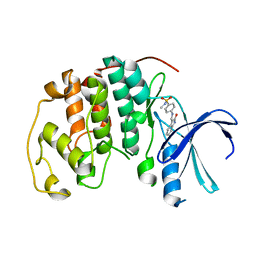 | | CDK2 complexed with N-methyl-4-{[(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]amino}benzenesulfonamide | | Descriptor: | Cell division protein kinase 2, N-METHYL-4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KE7
 
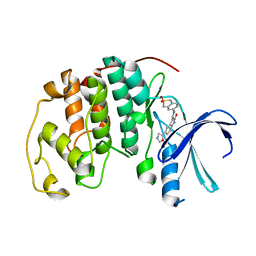 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 3-{[(2,2-DIOXIDO-1,3-DIHYDRO-2-BENZOTHIEN-5-YL)AMINO]METHYLENE}-5-(1,3-OXAZOL-5-YL)-1,3-DIHYDRO-2H-INDOL-2-ONE | | Descriptor: | 3-{[(2,2-DIOXIDO-1,3-DIHYDRO-2-BENZOTHIEN-5-YL)AMINO]METHYLENE}-5-(1,3-OXAZOL-5-YL)-1,3-DIHYDRO-2H-INDOL-2-ONE, Cell division protein kinase 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.H, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
2P8L
 
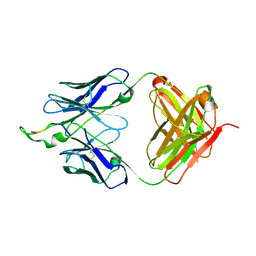 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELLELDKWASLWN | | Descriptor: | gp41 peptide, nmAb 2F5, heavy chain, ... | | Authors: | Julien, J.P, Bryson, S, Pai, E.F. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
1LXC
 
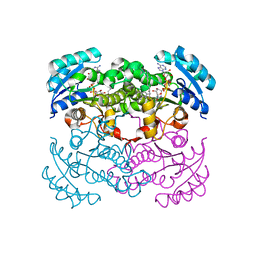 | | Crystal Structure of E. Coli Enoyl Reductase-NAD+ with a Bound Acrylamide Inhibitor | | Descriptor: | 3-(6-AMINOPYRIDIN-3-YL)-N-METHYL-N-[(1-METHYL-1H-INDOL-2-YL)METHYL]ACRYLAMIDE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Miller, W.H, Seefeld, M.A, Newlander, K.A, Uzinskas, I.N, Burgess, W.J, Heerding, D.A, Yuan, C.C.K, Head, M.S, Payne, D.J, Rittenhouse, S.F, Moore, T.D, Pearson, S.C, Dewolf, V, Berry, W.E, Keller, P.M, Polizzi, B.J, Qiu, X, Janson, C.A, Huffman, W.F. | | Deposit date: | 2002-06-05 | | Release date: | 2002-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of aminopyridine-based inhibitors of bacterial enoyl-ACP reductase (FabI).
J.Med.Chem., 45, 2002
|
|
1MFP
 
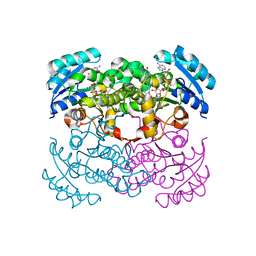 | | E. coli Enoyl Reductase in complex with NAD and SB611113 | | Descriptor: | (E)-N-METHYL-N-(1-METHYL-1H-INDOL-3-YLMETHYL)-3-(7-OXO-5,6,7,8-TETRAHYDRO-[1,8]NAPHTHYRIDIN-3-YL)-ACRYLAMIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Seefeld, M.A, Miller, W.H, Newlander, K.A, Burgess, W.J, DeWolf Jr, W.E, Elkins, P.A, Head, M.S, Jakas, D.R, Janson, C.A, Keller, P.M, Manley, P.J, Moore, T.D, Payne, D.J, Pearson, S, Polizzi, B.J, Qiu, X, Rittenhouse, S.F, Uzinskas, I.N, Wallis, N.G, Huffman, W.F. | | Deposit date: | 2002-08-13 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Indole Naphthyridinones as Inhibitors of Bacterial Enoyl-ACP Reductases FabI and FabK
J.MED.CHEM., 46, 2003
|
|
1HMV
 
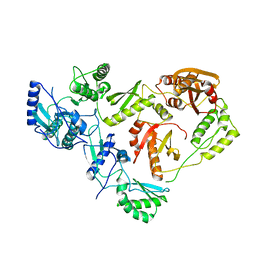 | | THE STRUCTURE OF UNLIGANDED REVERSE TRANSCRIPTASE FROM THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66), MAGNESIUM ION | | Authors: | Rodgers, D.W, Gamblin, S.J, Harris, B.A, Ray, S, Culp, J.S, Hellmig, B, Woolf, D.J, Debouck, C, Harrison, S.C. | | Deposit date: | 1994-12-15 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
2QQZ
 
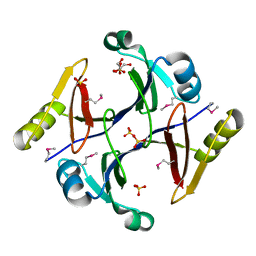 | | Crystal structure of putative glyoxalase family protein from Bacillus anthracis | | Descriptor: | GLYCEROL, Glyoxalase family protein, putative, ... | | Authors: | Kim, Y, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Putative Glyoxalase Family Protein from Bacillus anthracis.
To be Published
|
|
1Q9C
 
 | |
1YIL
 
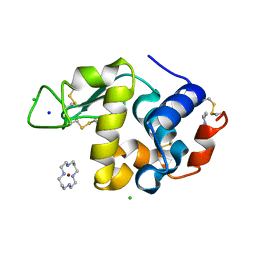 | | Structure of Hen egg white lysozyme soaked with Cu2-Xylylbicyclam | | Descriptor: | 1,1'-[1,4-PHENYLENEBIS(METHYLENE)]BIS[1,4,8,11-TETRAAZA-CYCLOTETRADECANE]CU(II)2, CHLORIDE ION, Lysozyme C, ... | | Authors: | Hunter, T.M, McNae, I.W, Liang, X, Bella, J, Parsons, S, Walkinshaw, M.D, Sadler, P.J. | | Deposit date: | 2005-01-12 | | Release date: | 2005-02-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein recognition of macrocycles: binding of anti-HIV metallocyclams to lysozyme
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
