2MFX
 
 | | Non-reducible analogues of alpha-conotoxin Vc1.1: [2,8]-cis dicarba Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Robinson, S.D, Macraild, C.A, Van Lierop, B.J, Robinson, A.J, Norton, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2013-12-18 | | Method: | SOLUTION NMR | | Cite: | Dicarba alpha-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors.
Acs Chem.Biol., 8, 2013
|
|
1G28
 
 | |
2WKB
 
 | | Crystal Structure of Macrophage Migration Inhibitory Factor from Plasmodium berghei | | Descriptor: | GLYCEROL, MACROPHAGE MIGRATION INHIBITORY FACTOR | | Authors: | Dobson, S.E, Augustijn, K.D, Brannigan, J.A, Dodson, E.J, Waters, A.P, Wilkinson, A.J. | | Deposit date: | 2009-06-08 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Crystal Structures of Macrophage Migration Inhibition Factor (Mif) from Plasmodium Falciparum and Plasmodium Berghei.
Protein Sci., 18, 2009
|
|
6F9C
 
 | |
8I5P
 
 | |
8I5Q
 
 | |
3Q6U
 
 | | Structure of the apo MET receptor kinase in the dually-phosphorylated, activated state | | Descriptor: | Hepatocyte growth factor receptor | | Authors: | Soisson, S.M, Rickert, K.W, Patel, S.B, Allison, T, Lumb, K.J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for selective small molecule kinase inhibition of activated c-Met.
J.Biol.Chem., 286, 2011
|
|
2J7K
 
 | | Crystal structure of the T84A mutant EF-G:GDPCP complex | | Descriptor: | ELONGATION FACTOR G, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Hansson, S, Logan, D.T. | | Deposit date: | 2006-10-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | New Insights Into the Role of the P-Loop Lysine: Implications from the Crystal Structure of a Mutant EF-G:Gdpcp Complex
To be Published
|
|
1SAP
 
 | |
2LN7
 
 | |
1ZA6
 
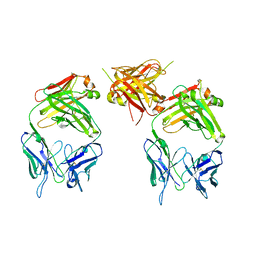 | | The structure of an antitumor CH2-domain-deleted humanized antibody | | Descriptor: | IGG Heavy chain, IGG Light chain | | Authors: | Larson, S.B, Day, J.S, Glaser, S, Braslawsky, G, McPherson, A. | | Deposit date: | 2005-04-05 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of an Antitumor C(H)2-domain-deleted Humanized Antibody.
J.Mol.Biol., 348, 2005
|
|
3A5F
 
 | |
1RFA
 
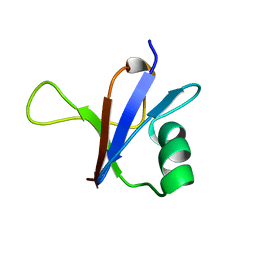 | | NMR SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF C-RAF-1 | | Descriptor: | RAF1 | | Authors: | Emerson, S.D, Madison, V.S, Palermo, R.E, Waugh, D.S, Scheffler, J.E, Tsao, K.-L, Kiefer, S.E, Liu, S.P, Fry, D.C. | | Deposit date: | 1995-04-26 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of c-Raf-1 and identification of its Ras interaction surface.
Biochemistry, 34, 1995
|
|
2LNJ
 
 | |
2KDS
 
 | |
3TY0
 
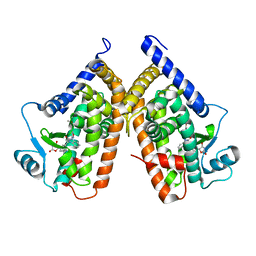 | | Structure of PPARgamma ligand binding domain in complex with (R)-5-(3-((3-(6-methoxybenzo[d]isoxazol-3-yl)-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)methyl)phenyl)-5-methyloxazolidine-2,4-dione | | Descriptor: | (5R)-5-(3-{[3-(6-methoxy-1,2-benzoxazol-3-yl)-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl]methyl}phenyl)-5-methyl-1,3-oxazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Soisson, S.M, Meinke, P.M, McKeever, B, Liu, W. | | Deposit date: | 2011-09-23 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Benzimidazolones: a new class of selective peroxisome proliferator-activated receptor gamma (PPAR-gamma) modulators.
J.Med.Chem., 54, 2011
|
|
8KC6
 
 | | X-ray structure of 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase | | Descriptor: | 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase | | Authors: | Song, S.K, Lv, D.Q, Cheng, X.Y, Ma, Q.Q, Hu, Y.L, Su, Z.D. | | Deposit date: | 2023-08-06 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray structure of 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase
To Be Published
|
|
3L2M
 
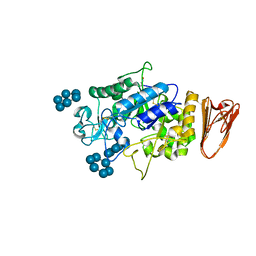 | | X-ray Crystallographic Analysis of Pig Pancreatic Alpha-Amylase with Alpha-cyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2009-12-15 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | X-ray crystallographic analyses of pig pancreatic alpha-amylase with limit dextrin, oligosaccharide, and alpha-cyclodextrin.
Biochemistry, 49, 2010
|
|
4OQ9
 
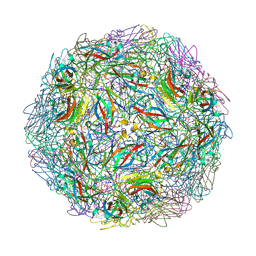 | | Satellite Tobacco Mosaic Virus Refined to 1.4 A Resolution using non-crystallographic symmetry restraints | | Descriptor: | Coat protein, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2014-02-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Satellite tobacco mosaic virus refined to 1.4 angstrom resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2F5A
 
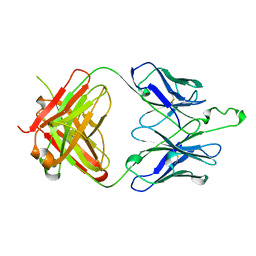 | | CRYSTAL STRUCTURE OF FAB' FROM THE HIV-1 NEUTRALIZING ANTIBODY 2F5 | | Descriptor: | PROTEIN (ANTIBODY 2F5 (HEAVY CHAIN)), PROTEIN (ANTIBODY 2F5 (LIGHT CHAIN)) | | Authors: | Bryson, S, Julien, J.P, Hynes, R.C, Pai, E.F. | | Deposit date: | 1999-04-08 | | Release date: | 2003-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications.
J.Virol., 83, 2009
|
|
4NIA
 
 | | Satellite Tobacco Mosaic Virus Refined at room temperature to 1.8 A Resolution using NCS Restraints | | Descriptor: | Coat protein, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Satellite tobacco mosaic virus refined to 1.4 angstrom resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8I5S
 
 | | Crystal structure of TxGH116 D593N acid/base mutant from Thermoanaerobacterium xylanolyticum with 2-deoxy-2-fluoroglucoside | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, 2-deoxy-2-fluoro-alpha-D-glucopyranose, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2023-01-26 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reaction Mechanism of Glycoside Hydrolase Family 116 Utilizes Perpendicular Protonation.
Acs Catalysis, 13, 2023
|
|
8I5T
 
 | |
8I5U
 
 | |
7DKV
 
 | |
