4Q57
 
 | | Crystal structure of the plectin 1a actin-binding domain/N-terminal domain of calmodulin complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Song, J.-G, Kostan, J, Grishkovskaya, I, Djinovic-Carugo, K. | | Deposit date: | 2014-04-16 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the plectin 1a actin-binding domain/N-terminal domain of calmodulin complex
To be Published
|
|
4NXS
 
 | | Crystal structure of human alpha-galactosidase A in complex with 1-deoxygalactonojirimycin-pFPhT | | Descriptor: | (2R,3S,4R,5S)-N-(4-fluorophenyl)-3,4,5-trihydroxy-2-(hydroxymethyl)piperidine-1-carbothioamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Johnson, J.L, Drury, J.E, Lieberman, R.L. | | Deposit date: | 2013-12-09 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5493 Å) | | Cite: | Molecular Basis of 1-Deoxygalactonojirimycin Arylthiourea Binding to Human alpha-Galactosidase A: Pharmacological Chaperoning Efficacy on Fabry Disease Mutants.
Acs Chem.Biol., 9, 2014
|
|
5CDY
 
 | |
1H3I
 
 | | Crystal structure of the Histone Methyltransferase SET7/9 | | Descriptor: | HISTONE H3 LYSINE 4 SPECIFIC METHYLTRANSFERASE, MAGNESIUM ION | | Authors: | Wilson, J.R, Jing, C, Walker, P.A, Martin, S.R, Howell, S.A, Blackburn, G.M, Gamblin, S.J, Xiao, B. | | Deposit date: | 2002-09-04 | | Release date: | 2002-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Functional Analysis of the Histone Methyltransferase Set7/9
Cell(Cambridge,Mass.), 111, 2002
|
|
5TDS
 
 | |
5TDU
 
 | |
4RYB
 
 | |
3GJ1
 
 | | Non photoactivated state of PA-GFP | | Descriptor: | CHLORIDE ION, Green fluorescent protein, SULFATE ION | | Authors: | Henderson, J.N, Gepshtein, R, Heenan, J.R, Kallio, K, Huppert, D, Remington, S.J. | | Deposit date: | 2009-03-07 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of the photoactivatable green fluorescent protein.
J.Am.Chem.Soc., 131, 2009
|
|
3UB7
 
 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with acetamide bound | | Descriptor: | ACETAMIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | Deposit date: | 2011-10-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
3UB8
 
 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with formamide bound | | Descriptor: | FORMAMIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | Deposit date: | 2011-10-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
3HOT
 
 | |
5FWG
 
 | | TETRA-(5-FLUOROTRYPTOPHANYL)-GLUTATHIONE TRANSFERASE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, TETRA-(5-FLUOROTRYPTOPHANYL)-GLUTATHIONE TRANSFERASE MU CLASS | | Authors: | Parsons, J.F, Xiao, G, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1997-11-08 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymes harboring unnatural amino acids: mechanistic and structural analysis of the enhanced catalytic activity of a glutathione transferase containing 5-fluorotryptophan.
Biochemistry, 37, 1998
|
|
4K07
 
 | | Crystal structure of the amyloid-forming immunoglobulin AL-103 cis-proline 95 mutant | | Descriptor: | Amyloidogenic immunoglobulin light chain protein AL-103, SULFATE ION | | Authors: | Thompson, J.R, Berkholz, D.S, Mahlum, E.W, Ramirez-Alvarado, M. | | Deposit date: | 2013-04-03 | | Release date: | 2013-10-30 | | Last modified: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Kinetic control in protein folding for light chain amyloidosis and the differential effects of somatic mutations.
J.Mol.Biol., 426, 2014
|
|
2A47
 
 | | Crystal structure of amFP486 H199T | | Descriptor: | BETA-MERCAPTOETHANOL, GFP-like fluorescent chromoprotein amFP486 | | Authors: | Henderson, J.N, Remington, S.J. | | Deposit date: | 2005-06-28 | | Release date: | 2005-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures and mutational analysis of amFP486, a cyan fluorescent protein from Anemonia majano
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3HOS
 
 | |
3IR8
 
 | | Red fluorescent protein mKeima at pH 7.0 | | Descriptor: | Large stokes shift fluorescent protein | | Authors: | Henderson, J.N, Osborn, M.F, Koon, N, Gepshtein, R, Huppert, D, Remington, S.J. | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Excited state proton transfer in the red fluorescent protein mKeima.
J.Am.Chem.Soc., 131, 2009
|
|
3UB9
 
 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with hydroxyurea bound | | Descriptor: | GLYCEROL, N-HYDROXYUREA, SULFATE ION, ... | | Authors: | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | Deposit date: | 2011-10-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
2A46
 
 | |
4TRD
 
 | |
4U7B
 
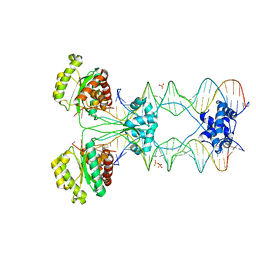 | | Crystal structure of a pre-cleavage Mos1 transpososome | | Descriptor: | DNA (25-MER), DNA (31-MER), Mariner Mos1 transposase, ... | | Authors: | Richardson, J.M. | | Deposit date: | 2014-07-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural role of the flanking DNA in mariner transposon excision.
Nucleic Acids Res., 43, 2015
|
|
3UB6
 
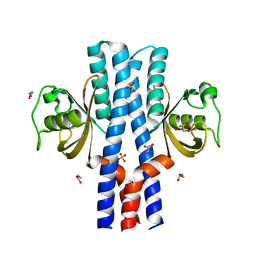 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with urea bound | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | Deposit date: | 2011-10-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
2A48
 
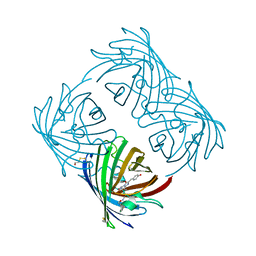 | | Crystal structure of amFP486 E150Q | | Descriptor: | BETA-MERCAPTOETHANOL, GFP-like fluorescent chromoprotein amFP486 | | Authors: | Henderson, J.N, Remington, S.J. | | Deposit date: | 2005-06-28 | | Release date: | 2005-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and mutational analysis of amFP486, a cyan fluorescent protein from Anemonia majano
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5UUM
 
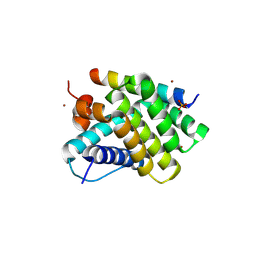 | | Human Mcl-1 in complex with a Bfl-1-specific selected peptide | | Descriptor: | Bfl-1 specific peptide FS2, Induced myeloid leukemia cell differentiation protein Mcl-1, SULFATE ION, ... | | Authors: | Jenson, J.M, Grant, R.A, Keating, A.E. | | Deposit date: | 2017-02-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | Epistatic mutations in PUMA BH3 drive an alternate binding mode to potently and selectively inhibit anti-apoptotic Bfl-1.
Elife, 6, 2017
|
|
2Y3R
 
 | | Structure of the tirandamycin-bound FAD-dependent tirandamycin oxidase TamL in P21 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-12-22 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
2Y3S
 
 | | Structure of the tirandamycine-bound FAD-dependent tirandamycin oxidase TamL in C2 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-12-23 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
