1HGE
 
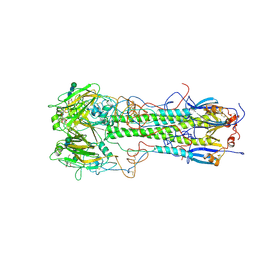 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
5W8F
 
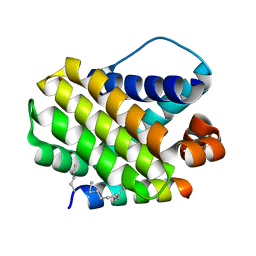 | | Crystal structure of human Mcl-1 in complex with modified Bim BH3 peptide SAH-MS1-14 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, modified Bim BH3 peptide SAH-MS1-14 | | Authors: | Rezaei Araghi, R, Jenson, J.M, Grant, R.A, Keating, A.E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Iterative optimization yields Mcl-1-targeting stapled peptides with selective cytotoxicity to Mcl-1-dependent cancer cells.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3PD7
 
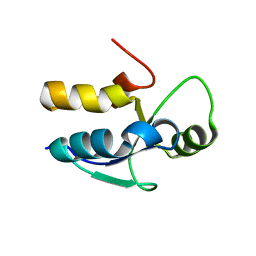 | |
1HGD
 
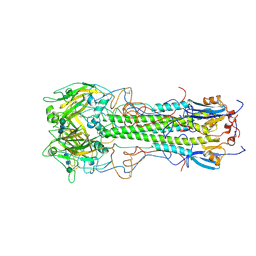 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
1MPL
 
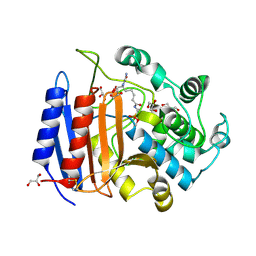 | | CRYSTAL STRUCTURE OF PHOSPHONATE-INHIBITED D-ALA-D-ALA PEPTIDASE REVEALS AN ANALOG OF A TETRAHEDRAL TRANSITION STATE | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, GLYCEROL, GLYCYL-L-A-AMINOPIMELYL-E-(D-2-AMINOETHYL)PHOSPHONATE | | Authors: | Silvaggi, N.R, Anderson, J.W, Brinsmade, S.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2002-09-12 | | Release date: | 2003-02-25 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The Crystal Structure of Phosphonate-Inhibited
d-Ala-d-Ala Peptidase Reveals an Analogue of a Tetrahedral Transition State.
Biochemistry, 42, 2003
|
|
3PEP
 
 | |
3RFH
 
 | |
3GSB
 
 | |
3RFG
 
 | |
4PL3
 
 | | Crystal structure of murine IRE1 in complex with MKC9989 inhibitor | | Descriptor: | 7-hydroxy-6-methoxy-3-[2-(2-methoxyethoxy)ethyl]-4,8-dimethyl-2H-chromen-2-one, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sanches, M, Duffy, N, Talukdar, M, Thevakumaran, N, Chiovitti, D, Al-awar, R, Patterson, J.B, Sicheri, F. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of action of the hydroxy-aryl-aldehyde class of IRE1 endoribonuclease inhibitors.
Nat Commun, 5, 2014
|
|
3QPR
 
 | | HK97 Prohead I encapsidating inactive virally encoded protease | | Descriptor: | Major capsid protein | | Authors: | Huang, R.K, Khayat, R, Lee, K.K, Gertsman, I, Duda, R.L, Hendrix, R.W, Johnson, J.E. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.2 Å) | | Cite: | The Prohead-I structure of bacteriophage HK97: implications for scaffold-mediated control of particle assembly and maturation.
J.Mol.Biol., 408, 2011
|
|
4L6S
 
 | | PARP complexed with benzo[1,4]oxazin-3-one inhibitor | | Descriptor: | (2S)-6-{[4-(4-chlorophenyl)-3,6-dihydropyridin-1(2H)-yl]methyl}-2-methyl-2H-1,4-benzoxazin-3(4H)-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Dougan, D.R, Mol, C.D, Lawson, J.D. | | Deposit date: | 2013-06-12 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of novel benzo[b][1,4]oxazin-3(4H)-ones as poly(ADP-ribose)polymerase inhibitors
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3QJC
 
 | |
3QJB
 
 | |
3RF2
 
 | |
4MQC
 
 | |
5V3T
 
 | | Crystal Structure of the Group II Truncated Hemoglobin from Bacillus Anthracis | | Descriptor: | CYANIDE ION, Globin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Varnado, C.L, SoRelle, E, Soman, J, Olson, J.S. | | Deposit date: | 2017-03-08 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Crystal Structure of the Group II Truncated Hemoglobin from Bacillus Anthracis
To Be Published
|
|
4MQH
 
 | |
5V3V
 
 | | Crystal Structure of the Group II Truncated Hemoglobin from Bacillus Anthracis: Tyr26Ala Mutant | | Descriptor: | CYANIDE ION, Globin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Varnado, C.L, SoRelle, E, Soman, J, Olson, J.S. | | Deposit date: | 2017-03-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal Structure of the Group II Truncated Hemoglobin from Bacillus Anthracis
To Be Published
|
|
2IP2
 
 | |
3OTI
 
 | | Crystal Structure of CalG3, Calicheamicin Glycostyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CHLORIDE ION, CalG3, Calicheamicin T0, ... | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2010-09-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QJE
 
 | |
2F12
 
 | | Crystal Structure of the Human Sialidase Neu2 in Complex with 3- hydroxypropyl ether mimetic Inhibitor | | Descriptor: | 3-hydroxypropyl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Mann, M.C, Thomson, R.J, Dyason, J.C, von Itzstein, M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with 3- hydroxypropyl ether mimetic Inhibitor
To be Published
|
|
1GC1
 
 | | HIV-1 GP120 CORE COMPLEXED WITH CD4 AND A NEUTRALIZING HUMAN ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, CD4, ... | | Authors: | Kwong, P.D, Wyatt, R, Robinson, J, Sweet, R.W, Sodroski, J, Hendrickson, W.A. | | Deposit date: | 1998-06-15 | | Release date: | 1998-07-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody.
Nature, 393, 1998
|
|
5TLQ
 
 | | Model structure of the oxidized PaDsbA1 and 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine complex | | Descriptor: | 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, Thiol:disulfide interchange protein DsbA | | Authors: | Mohanty, B, Rimmer, K.A, McMahon, R.M, Headey, S.J, Vazirani, M, Shouldice, S.R, Coincon, M, Tay, S, Morton, C.J, Simpson, J.S, Martin, J.L, Scanlon, M.S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
