4J40
 
 | | Crystal structure of the dual-domain GGDEF-EAL module of FimX from Pseudomonas aeruginosa | | Descriptor: | FimX | | Authors: | Navarro, M.V, De, N, Bae, N, Wang, Q, Sondermann, H. | | Deposit date: | 2013-02-06 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX.
Structure, 17, 2009
|
|
2KA4
 
 | | NMR structure of the CBP-TAZ1/STAT2-TAD complex | | Descriptor: | Crebbp protein, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains
Embo J., 28, 2009
|
|
1Q67
 
 | | Crystal structure of Dcp1p | | Descriptor: | Decapping protein involved in mRNA degradation-Dcp1p | | Authors: | She, M, Decker, C.J, Liu, Y, Chen, N, Parker, R, Song, H. | | Deposit date: | 2003-08-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Dcp1p and its functional implications in mRNA decapping
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RXR
 
 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE RETINOID X RECEPTOR DNA BINDING DOMAIN, NMR, 20 STRUCTURE | | Descriptor: | RETINOIC ACID RECEPTOR-ALPHA, ZINC ION | | Authors: | Holmbeck, S.M.A, Foster, M.P, Casimiro, D.R, Sem, D.S, Dyson, H.J, Wright, P.E. | | Deposit date: | 1998-06-12 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the retinoid X receptor DNA-binding domain.
J.Mol.Biol., 281, 1998
|
|
1R5O
 
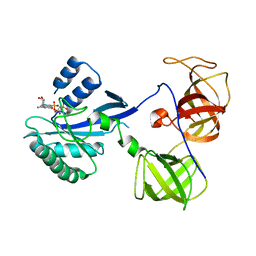 | | crystal structure analysis of sup35 complexed with GMPPNP | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-11 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
3Q5D
 
 | |
2KA6
 
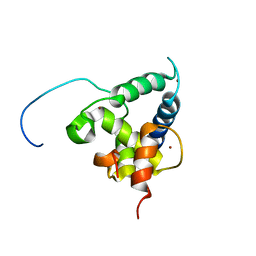 | | NMR structure of the CBP-TAZ2/STAT1-TAD complex | | Descriptor: | CREB-binding protein, Signal transducer and activator of transcription 1-alpha/beta, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains.
Embo J., 28, 2009
|
|
4IZA
 
 | |
1SB0
 
 | | Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb | | Descriptor: | protein CBP, protein c-Myb | | Authors: | Zor, T, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-02-09 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the KIX Domain of CBP Bound to the Transactivation
Domain of c-Myb
J.Mol.Biol., 337, 2004
|
|
2H8G
 
 | | 5'-Methylthioadenosine Nucleosidase from Arabidopsis thaliana | | Descriptor: | 5'-Methylthioadenosine Nucleosidase, ADENINE | | Authors: | Park, E.Y, Oh, S.I, Nam, M.J, Shin, J.S, Kim, K.N, Song, H.K. | | Deposit date: | 2006-06-07 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine nucleosidase from Arabidopsis thaliana at 1.5-A resolution
Proteins, 65, 2006
|
|
4IZ7
 
 | |
4JX7
 
 | | Crystal structure of Pim1 kinase in complex with inhibitor 2-[(trans-4-aminocyclohexyl)amino]-4-{[3-(trifluoromethyl)phenyl]amino}pyrido[4,3-d]pyrimidin-5(6H)-one | | Descriptor: | 2-[(trans-4-aminocyclohexyl)amino]-4-{[3-(trifluoromethyl)phenyl]amino}pyrido[4,3-d]pyrimidin-5(6H)-one, PIM1 consensus peptide, Serine/threonine-protein kinase pim-1 | | Authors: | Lee, S.J, Han, B.G, Cho, J.W, Choi, J.S, Lee, J.K, Song, H.J, Koh, J.S, Lee, B.I. | | Deposit date: | 2013-03-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of pim1 kinase in complex with a pyrido[4,3-d]pyrimidine derivative suggests a unique binding mode.
Plos One, 8, 2013
|
|
1MNV
 
 | | Actinomycin D binding to ATGCTGCAT | | Descriptor: | 5'-D(*AP*TP*GP*CP*TP*GP*CP*AP*T)-3', ACTINOMYCIN D | | Authors: | Hou, M.-H, Robinson, H, Gao, Y.-G, Wang, A.H.-J. | | Deposit date: | 2002-09-06 | | Release date: | 2002-11-22 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Actinomycin D Bound to the Ctg Triplet Repeat Sequences Linked to Neurological Diseases
Nucleic Acids Res., 30, 2002
|
|
2HDP
 
 | | Solution Structure of Hdm2 RING Finger Domain | | Descriptor: | Ubiquitin-protein ligase E3 Mdm2, ZINC ION | | Authors: | Kostic, M, Matt, T, Yamout-Martinez, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2006-06-20 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53.
J.Mol.Biol., 363, 2006
|
|
2GJK
 
 | | Structural and functional insights into the human Upf1 helicase core | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Regulator of nonsense transcripts 1 | | Authors: | Cheng, Z, Muhlrad, D, Parker, R, Song, H. | | Deposit date: | 2006-03-31 | | Release date: | 2007-01-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insights into the human Upf1 helicase core
Embo J., 26, 2007
|
|
2GK7
 
 | | Structural and Functional insights into the human Upf1 helicase core | | Descriptor: | PHOSPHATE ION, Regulator of nonsense transcripts 1 | | Authors: | Cheng, Z, Muhlrad, D, Parker, R, Song, H. | | Deposit date: | 2006-03-31 | | Release date: | 2007-01-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional insights into the human Upf1 helicase core
Embo J., 26, 2007
|
|
1DGS
 
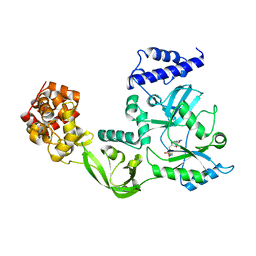 | | CRYSTAL STRUCTURE OF NAD+-DEPENDENT DNA LIGASE FROM T. FILIFORMIS | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA LIGASE, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Kwon, S.T, Suh, S.W. | | Deposit date: | 1999-11-25 | | Release date: | 2000-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
EMBO J., 19, 2000
|
|
2L14
 
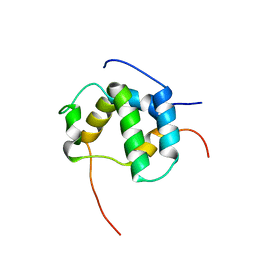 | | Structure of CBP nuclear coactivator binding domain in complex with p53 TAD | | Descriptor: | CREB-binding protein, Cellular tumor antigen p53 | | Authors: | Lee, C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2010-07-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein.
Biochemistry, 49, 2010
|
|
2FJT
 
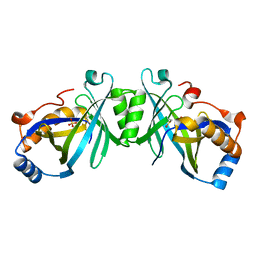 | | Adenylyl cyclase class iv from Yersinia pestis | | Descriptor: | Adenylyl cyclase class IV, SULFATE ION | | Authors: | Gallagher, D.T, Smith, N.N, Kim, S.-K, Reddy, P.T, Robinson, H, Heroux, A. | | Deposit date: | 2006-01-03 | | Release date: | 2006-11-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of the class IV adenylyl cyclase reveals a novel fold
J.Mol.Biol., 362, 2006
|
|
1U2N
 
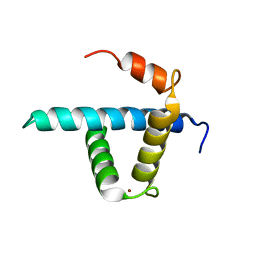 | | Structure CBP TAZ1 Domain | | Descriptor: | CREB binding protein, ZINC ION | | Authors: | De Guzman, R.N, Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-07-19 | | Release date: | 2005-04-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding
Biochemistry, 44, 2005
|
|
1R8U
 
 | | NMR structure of CBP TAZ1/CITED2 complex | | Descriptor: | CREB-binding protein, Cbp/p300-interacting transactivator 2, ZINC ION | | Authors: | De Guzman, R.N, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-10-28 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2: regulation by competition between intrinsically unstructured ligands for non-identical binding sites.
J.Biol.Chem., 279, 2004
|
|
2QKM
 
 | |
7LVS
 
 | | The CBP TAZ1 Domain in Complex with a CITED2-HIF-1-Alpha Fusion Peptide | | Descriptor: | Cbp/p300-interacting transactivator 2,Hypoxia-inducible factor 1-alpha, Histone lysine acetyltransferase CREBBP, ZINC ION | | Authors: | Appling, F.D, Berlow, R.B, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The molecular basis of allostery in a facilitated dissociation process.
Structure, 29, 2021
|
|
1C76
 
 | | STAPHYLOKINASE (SAK) MONOMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Staphylokinase Dimer Offers New Clue to Reduction of Immunogenicity
To be published
|
|
1KBH
 
 | | Mutual Synergistic Folding in the Interaction Between Nuclear Receptor Coactivators CBP and ACTR | | Descriptor: | CREB-BINDING PROTEIN, nuclear receptor coactivator | | Authors: | Demarest, S.J, Martinez-Yamout, M, Chung, J, Chen, H, Xu, W, Dyson, H.J, Evans, R.M, Wright, P.E. | | Deposit date: | 2001-11-06 | | Release date: | 2002-02-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators.
Nature, 415, 2002
|
|
