1JOD
 
 | |
1JOB
 
 | |
4FIN
 
 | | Crystal Structure of EttA (formerly YjjK) - an E. coli ABC-type ATPase | | Descriptor: | CITRIC ACID, EttA (YjjK) ABCF family protein, GLYCEROL, ... | | Authors: | Smith, P, Yuan, Y, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-09 | | Release date: | 2013-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The ABC-F protein EttA gates ribosome entry into the translation elongation cycle.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6S54
 
 | | Transaminase from Pseudomonas fluorescens | | Descriptor: | Aspartate aminotransferase family protein, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Smith, P, Roura Padrosa, D, Lopez-Gallego, F, Paradisi, F, Dreveny, I. | | Deposit date: | 2019-06-30 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Enhancing PLP-Binding Capacity of Class-III omega-Transaminase by Single Residue Substitution.
Front Bioeng Biotechnol, 7, 2019
|
|
3TY8
 
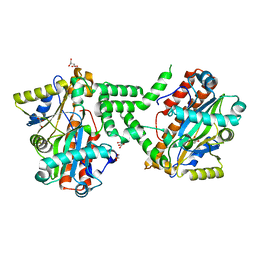 | | Crystal Structure of C. Thermocellum PNKP Ligase Domain Apo Form | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Polynucleotide 2',3'-cyclic phosphate phosphodiesterase / polynucleotide 5'-hydroxyl-kinase / polynucleotide 3'-phosphatase | | Authors: | Smith, P, Wang, L, Shuman, S. | | Deposit date: | 2011-09-23 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The adenylyltransferase domain of bacterial Pnkp defines a unique RNA ligase family.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TY5
 
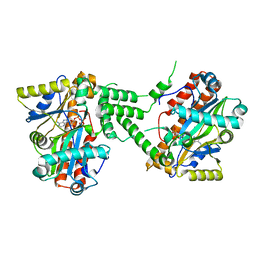 | | Crystal Structure of C. thermocellum PNKP Ligase domain in complex with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Polynucleotide 2',3'-cyclic phosphate phosphodiesterase / polynucleotide 5'-hydroxyl-kinase / polynucleotide 3'-phosphatase, ... | | Authors: | Smith, P, Wang, L, Shuman, S. | | Deposit date: | 2011-09-23 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The adenylyltransferase domain of bacterial Pnkp defines a unique RNA ligase family.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TY9
 
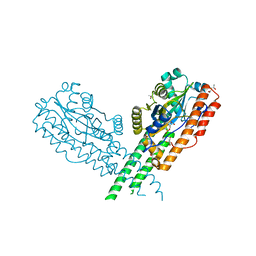 | | Crystal Structure of C. Thermocellum PNKP Ligase Domain AMP-Adenylate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Smith, P, Wang, L, Shuman, S. | | Deposit date: | 2011-09-24 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The adenylyltransferase domain of bacterial Pnkp defines a unique RNA ligase family.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4KYY
 
 | |
1L2T
 
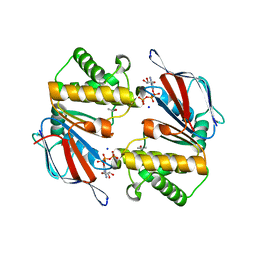 | | Dimeric Structure of MJ0796, a Bacterial ABC Transporter Cassette | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Hypothetical ABC transporter ATP-binding protein MJ0796, ISOPROPYL ALCOHOL, ... | | Authors: | Smith, P.C, Karpowich, N, Rosen, J, Hunt, J.F. | | Deposit date: | 2002-02-24 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer.
Mol.Cell, 10, 2002
|
|
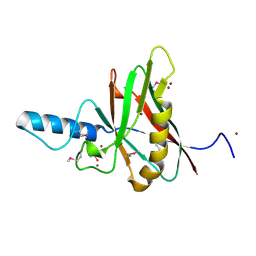
1F35
 
 | |
3HF1
 
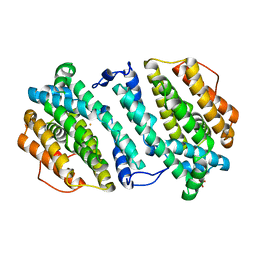 | | Crystal structure of human p53R2 | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase subunit M2 B, SULFATE ION | | Authors: | Smith, P, Zhou, B, Yuan, Y.-C, Su, L, Tsai, S.-C, Yen, Y. | | Deposit date: | 2009-05-10 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 A X-ray crystal structure of human p53R2, a p53-inducible ribonucleotide reductase .
Biochemistry, 48, 2009
|
|
3P43
 
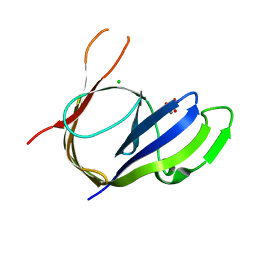 | | Structure and Activities of Archaeal Members of the LigD 3' Phosphoesterase DNA Repair Enzyme Superfamily | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Smith, P, Nair, P.A, Das, U, Shuman, S. | | Deposit date: | 2010-10-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures and activities of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily.
Nucleic Acids Res., 39, 2011
|
|
3P4H
 
 | | Structures of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily | | Descriptor: | ATP-dependent DNA ligase, N-terminal domain protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Smith, P, Nair, P.A, Das, U, Zhu, H, Shuman, S. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures and activities of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily.
Nucleic Acids Res., 39, 2011
|
|
3BGY
 
 | |
4W7S
 
 | | Crystal structure of the yeast DEAD-box splicing factor Prp28 at 2.54 Angstroms resolution | | Descriptor: | GLYCEROL, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Jacewicz, A, Smith, P, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Crystal structure, mutational analysis and RNA-dependent ATPase activity of the yeast DEAD-box pre-mRNA splicing factor Prp28.
Nucleic Acids Res., 42, 2014
|
|
5FQV
 
 | | Selective estrogen receptor downregulator antagonists: Tetrahydroisoquinoline phenols 5. | | Descriptor: | (E)-3-[4-(6-hydroxy-2-isobutyl-7-methyl-3,4-dihydro-1H-isoquinolin-1-yl)phenyl]prop-2-enoic acid, ESTROGEN RECEPTOR ALPHA | | Authors: | Scott, J.S, bailey, A, Davies, R.D.M, Degorce, S.L, MacFaul, P.A, Gingell, H, Moss, T, Norman, R.A, Pink, J.H, Rabow, A.A, Roberts, B, Smith, P.D. | | Deposit date: | 2015-12-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Tetrahydroisoquinoline Phenols: Selective Estrogen Receptor Downregulator Antagonists with Oral Bioavailability in Rat.
Acs Med.Chem.Lett., 7, 2016
|
|
3TA5
 
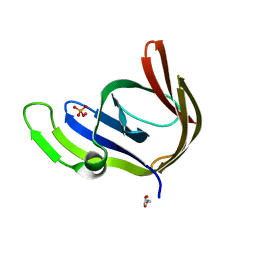 | | Cobalt bound structure of an archaeal member of the LigD 3'-phosphoesterase DNA repair enzyme family | | Descriptor: | ATP-dependent DNA ligase, N-terminal domain protein, COBALT (II) ION, ... | | Authors: | Das, U, Smith, P, Shuman, S. | | Deposit date: | 2011-08-03 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural insights to the metal specificity of an archaeal member of the LigD 3'-phosphoesterase DNA repair enzyme family.
Nucleic Acids Res., 40, 2012
|
|
3TA7
 
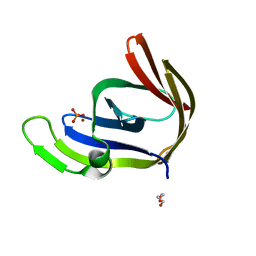 | | Zinc bound structure of an archaeal member of the LigD 3'-phosphoesterase DNA repair enzyme family | | Descriptor: | ATP-dependent DNA ligase, N-terminal domain protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Das, U, Smith, P, Shuman, S. | | Deposit date: | 2011-08-03 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural insights to the metal specificity of an archaeal member of the LigD 3'-phosphoesterase DNA repair enzyme family.
Nucleic Acids Res., 40, 2012
|
|
2XHD
 
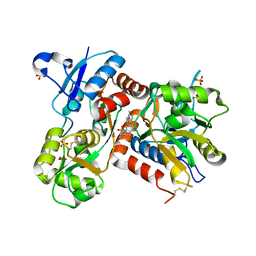 | | Crystal structure of N-((2S)-5-(6-fluoro-3-pyridinyl)-2,3-dihydro-1H- inden-2-yl)-2-propanesulfonamide in complex with the ligand binding domain of the human GluA2 receptor | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N-[(2S)-5-(6-FLUORO-3-PYRIDINYL)-2,3-DIHYDRO-1H-INDEN-2-YL]-2-PROPANESULFONAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Andreotti, D, Ballantine, S, Bax, B.D, Harris, A.J, Harker, A.J, Lund, J, Melarange, R, Mingardi, A, Mookherjee, C, Mosley, J, Neve, M, Oliosi, B, Profeta, R, Smith, K.J, Smith, P.W, Spada, S, Thewlis, K.M, Yusaf, S.P. | | Deposit date: | 2010-06-14 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of N-[(2S)-5-(6-Fluoro-3-Pyridinyl)-2,3-Dihydro-1H-Inden-2-Yl]-2-Propanesulfonamide, a Novel Clinical Ampa Receptor Positive Modulator.
J.Med.Chem., 53, 2010
|
|
4WAN
 
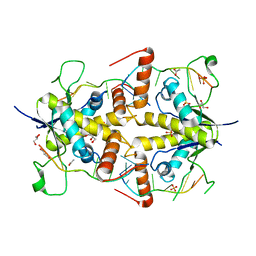 | | Crystal structure of Msl5 protein in complex with RNA at 1.8 A | | Descriptor: | ACETATE ION, Branchpoint-bridging protein, GLYCEROL, ... | | Authors: | Jacewicz, A, Smith, P, Chico, L, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs.
Rna, 21, 2015
|
|
4WAL
 
 | | Crystal structure of selenomethionine Msl5 protein in complex with RNA at 2.2 A | | Descriptor: | Branchpoint-bridging protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jacewicz, A, Smith, P, Chico, L, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs.
Rna, 21, 2015
|
|
7AVY
 
 | | MerTK kinase domain in complex with quinazoline-based inhbitor | | Descriptor: | N-(2-(2-cyclopropylethoxy)pyrimidin-5-yl)-7-methoxy-6-(piperidin-4-ylmethoxy)quinazolin-4-amine, SULFATE ION, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AVX
 
 | | MerTK kinase domain in complex with NPS-1034 | | Descriptor: | 1-(4-fluorophenyl)-N-[3-fluoro-4-[(3-phenyl-1H-pyrrolo[2,3-b]pyridin-4-yl)oxy]phenyl]-2,3-dimethyl-5-oxopyrazole-4-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW2
 
 | | MerTK kinase domain with type 1.5 inhibitor from a DNA-encoded library | | Descriptor: | 5-(2'-chloro-[1,1'-biphenyl]-4-yl)-N-(imidazo[1,2-a]pyridin-6-ylmethyl)-N-methyl-1,3,4-oxadiazol-2-amine, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW4
 
 | | MerTK kinase domain with type 3 inhibitor from a DNA-encoded library | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-5-(4-methyl-1,2,3-thiadiazol-5-yl)-N-((R)-1-(((R)-3-(methylamino)-3-oxo-1-(4-(trifluoromethyl)phenyl)propyl)amino)-1-oxo-4-phenylbutan-2-yl)isoxazole-4-carboxamide, CHLORIDE ION, ... | | Authors: | Pflug, A, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Preston, M, Rawlins, P, Rivers, E, Schimpl, M, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
