5WGT
 
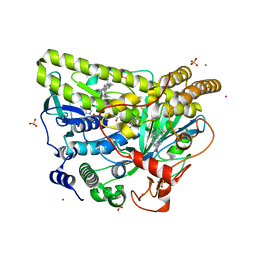 | | Crystal Structure of MalA' H253A, premalbrancheamide complex | | Descriptor: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
6ECW
 
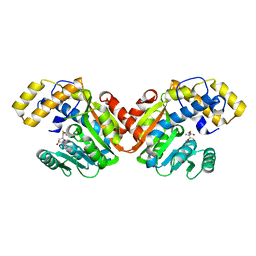 | | StiD O-MT residues 956-1266 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, StiD protein | | Authors: | Skiba, M.A, Bivins, M.M, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
5WM4
 
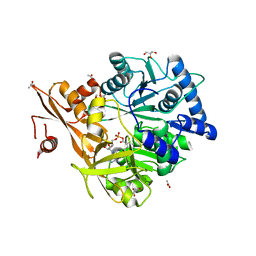 | | Crystal Structure of CahJ in Complex with 6-Methylsalicyl Adenylate | | Descriptor: | 9-(5-O-{(S)-hydroxy[(2-hydroxy-6-methylbenzene-1-carbonyl)oxy]phosphoryl}-alpha-L-lyxofuranosyl)-9H-purin-6-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Sikkema, A.P, Smith, J.L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ.
Chembiochem, 19, 2018
|
|
5W7K
 
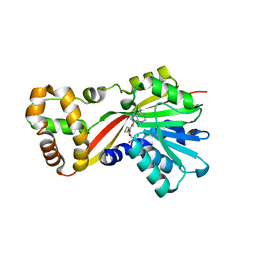 | | Crystal structure of OxaG | | Descriptor: | CHLORIDE ION, OxaG, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
5W7S
 
 | | Crystal structure of OxaC in complex with sinefungin and meleagrin | | Descriptor: | (3E,7aR,12aS)-6-hydroxy-3-[(1H-imidazol-4-yl)methylidene]-12-methoxy-7a-(2-methylbut-3-en-2-yl)-7a,12-dihydro-1H,5H-imidazo[1',2':1,2]pyrido[2,3-b]indole-2,5(3H)-dione, OxaC, SINEFUNGIN | | Authors: | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.948 Å) | | Cite: | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
5WM2
 
 | |
1NON
 
 | | PyrR, the regulator of the pyrimidine biosynthetic operon in Bacillus caldolyticus | | Descriptor: | PyrR bifunctional protein | | Authors: | Switzer, R.L, Chander, P, Smith, J.L, Halbig, K.M, Miller, J.K, Bonner, H.K, Grabner, G.K. | | Deposit date: | 2003-01-16 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Nucleotide Complex of PyrR, the pyr Attenuation Protein from Bacillus caldolyticus, Suggests Dual Regulation by Pyrimidine and Purine Nucleotides
J.Bacteriol., 187, 2005
|
|
2HFJ
 
 | | Pikromycin thioesterase with covalent pentaketide affinity label | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Akey, D.L, Kittendorf, J.D, Giraldes, J.W, Fecik, R.A, Sherman, D.H, Smith, J.L. | | Deposit date: | 2006-06-24 | | Release date: | 2006-09-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Macrolactonization by the Pikromycin Thioesterase
NAT.CHEM.BIOL., 2, 2006
|
|
1O57
 
 | | CRYSTAL STRUCTURE OF THE PURINE OPERON REPRESSOR OF BACILLUS SUBTILIS | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEXAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, ... | | Authors: | Sinha, S.C, Krahn, J, Shin, B.S, Tomchick, D.R, Zalkin, H, Smith, J.L. | | Deposit date: | 2003-04-20 | | Release date: | 2003-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Purine Repressor of Bacillus Subtilis: A Novel Combination of Domains Adapted for Transcription Regulation
J.Bacteriol., 185, 2003
|
|
2IGB
 
 | | Crystal Structure of PyrR, The Regulator Of The Pyrimidine Biosynthetic Operon In Bacillus caldolyticus, UMP-bound form | | Descriptor: | 1,2-ETHANEDIOL, PyrR bifunctional protein, URIDINE-5'-MONOPHOSPHATE | | Authors: | Chander, P, Switzer, R.L, Smith, J.L. | | Deposit date: | 2006-09-22 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PyrR, the regulator of the pyrimidine biosynthetic operon in Bacillus caldolyticus
To be Published
|
|
1P4A
 
 | | Crystal Structure of the PurR complexed with cPRPP | | Descriptor: | 1-ALPHA-PYROPHOSPHORYL-2-ALPHA,3-ALPHA-DIHYDROXY-4-BETA-CYCLOPENTANE-METHANOL-5-PHOSPHATE, Pur operon repressor | | Authors: | Bera, A.K, Zhu, J, Zalkin, H, Smith, J.L. | | Deposit date: | 2003-04-22 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Functional dissection of the Bacillus subtilis pur operator site.
J.Bacteriol., 185, 2003
|
|
2HFK
 
 | | Pikromycin thioesterase in complex with product 10-deoxymethynolide | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ETHYL-4-HYDROXY-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Akey, D.L, Kittendorf, J.D, Giraldes, J.W, Fecik, R.A, Sherman, D.H, Smith, J.L. | | Deposit date: | 2006-06-24 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Macrolactonization by the Pikromycin Thioesterase
NAT.CHEM.BIOL., 2, 2006
|
|
1ECJ
 
 | |
1A4X
 
 | | PYRR, THE BACILLUS SUBTILIS PYRIMIDINE BIOSYNTHETIC OPERON REPRESSOR, HEXAMERIC FORM | | Descriptor: | PYRIMIDINE OPERON REGULATORY PROTEIN PYRR, SULFATE ION | | Authors: | Tomchick, D.R, Turner, R.J, Switzer, R.W, Smith, J.L. | | Deposit date: | 1998-02-08 | | Release date: | 1998-08-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase.
Structure, 6, 1998
|
|
1AO0
 
 | |
1A3C
 
 | | PYRR, THE BACILLUS SUBTILIS PYRIMIDINE BIOSYNTHETIC OPERON REPRESSOR, DIMERIC FORM | | Descriptor: | PYRIMIDINE OPERON REGULATORY PROTEIN PYRR, SAMARIUM (III) ION, SULFATE ION | | Authors: | Tomchick, D.R, Turner, R.J, Switzer, R.W, Smith, J.L. | | Deposit date: | 1998-01-20 | | Release date: | 1998-08-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase.
Structure, 6, 1998
|
|
1ECC
 
 | |
1ECB
 
 | |
1E2V
 
 | | N153Q mutant of cytochrome f from Chlamydomonas reinhardtii | | Descriptor: | ACETATE ION, CYTOCHROME F, HEME C | | Authors: | Sainz, G, Carrell, C.J, Ponamarev, M.V, Soriano, G.M, Cramer, W.A, Smith, J.L. | | Deposit date: | 2000-05-29 | | Release date: | 2000-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Interruption of the Internal Water Chain of Cytochrome F Impairs Photosynthetic Function
Biochemistry, 39, 2000
|
|
1E2Z
 
 | | Q158L mutant of cytochrome f from Chlamydomonas reinhardtii | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Sainz, G, Carrell, C.J, Ponamarev, M.V, Soriano, G.M, Cramer, W.A, Smith, J.L. | | Deposit date: | 2000-05-30 | | Release date: | 2000-08-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interruption of the Internal Water Chain of Cytochrome F Impairs Photosynthetic Function
Biochemistry, 39, 2000
|
|
1E2W
 
 | | N168F mutant of cytochrome f from Chlamydomonas reinhardtii | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Sainz, G, Carrell, C.J, Ponamarev, M.V, Soriano, G.M, Cramer, W.A, Smith, J.L. | | Deposit date: | 2000-05-30 | | Release date: | 2000-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interruption of the Internal Water Chain of Cytochrome F Impairs Photosynthetic Function
Biochemistry, 39, 2000
|
|
1EWH
 
 | | STRUCTURE OF CYTOCHROME F FROM CHLAMYDOMONAS REINHARDTII | | Descriptor: | ACETATE ION, CYTOCHROME F, HEME C | | Authors: | Sainz, G, Carrell, C.J, Ponamarev, M.V, Soriano, G.M, Cramer, W.A, Smith, J.L. | | Deposit date: | 2000-04-25 | | Release date: | 2000-08-09 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Interruption of the internal water chain of cytochrome f impairs photosynthetic function.
Biochemistry, 39, 2000
|
|
3QIT
 
 | |
7K93
 
 | |
6MFD
 
 | | GphF GNAT-like decarboxylase in complex with isobutyryl-CoA | | Descriptor: | ACETATE ION, GLYCEROL, GphF, ... | | Authors: | Skiba, M.A, Tran, C.L, Smith, J.L. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Repurposing the GNAT Fold in the Initiation of Polyketide Biosynthesis.
Structure, 28, 2020
|
|
