3BOL
 
 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima complexed with Zn2+ | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, POTASSIUM ION, ... | | Authors: | Koutmos, M, Smith, J.L, Ludwig, M.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BQ6
 
 | |
3BQ5
 
 | | Crystal Structure of T. maritima Cobalamin-Independent Methionine Synthase complexed with Zn2+ and Homocysteine (Monoclinic) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase, ... | | Authors: | Pejchal, R, Smith, J.L, Ludwig, M.L. | | Deposit date: | 2007-12-19 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Q34
 
 | |
2Q35
 
 | |
2Q2X
 
 | |
8G7X
 
 | | Human fatty acid synthase dehydratase domain | | Descriptor: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Akey, D.L, Konwerski, J.R, McCullough, T.M, Smith, J.L. | | Deposit date: | 2023-02-17 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.815 Å) | | Cite: | Structure of a modular polyketide synthase reducing region.
Structure, 31, 2023
|
|
8G7W
 
 | | Type I modPKS reducing region | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | McCullough, T.M, Smith, J.L. | | Deposit date: | 2023-02-17 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a modular polyketide synthase reducing region.
Structure, 31, 2023
|
|
6ECX
 
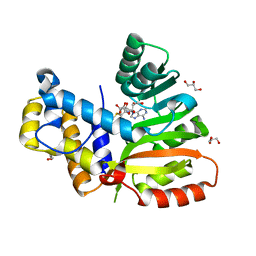 | | StiE O-MT residues 942-1257 | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, StiE protein | | Authors: | Skiba, M.A, Bivins, M.B, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
6ECV
 
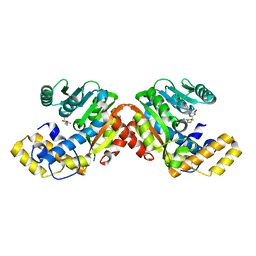 | | StiD O-MT residues 976-1266 | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, StiD protein | | Authors: | Skiba, M.A, Bivins, M.M, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
6ECT
 
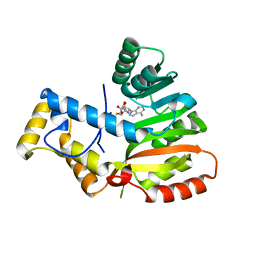 | | StiE O-MT residues 961-1257 | | Descriptor: | S-ADENOSYLMETHIONINE, StiE protein | | Authors: | Skiba, M.A, Bivins, M.M, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
6ECU
 
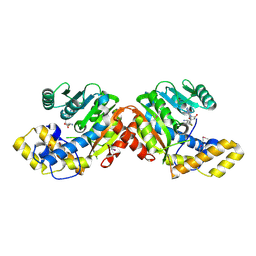 | |
5WGV
 
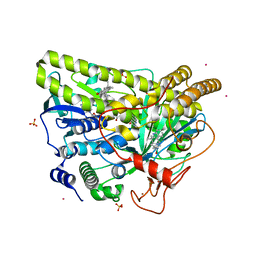 | | Crystal Structure of MalA' C112S/C128S, premalbrancheamide complex | | Descriptor: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5W7M
 
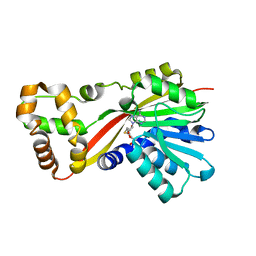 | | Crystal structure of RoqN | | Descriptor: | Glandicoline B O-methyltransferase roqN, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
5WGU
 
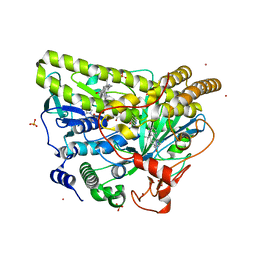 | | Crystal Structure of MalA' E494D, premalbrancheamide complex | | Descriptor: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WGS
 
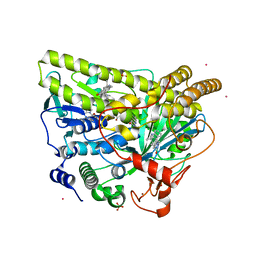 | | Crystal Structure of MalA' H253F, premalbrancheamide complex | | Descriptor: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WGR
 
 | | Crystal Structure of Wild-type MalA', premalbrancheamide complex | | Descriptor: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.362 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WGZ
 
 | | Crystal Structure of Wild-type MalA', isomalbrancheamide B complex | | Descriptor: | (5aS,12aS,13aS)-8-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WGX
 
 | | Crystal Structure of MalA' H253A, malbrancheamide B complex | | Descriptor: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WM3
 
 | | Crystal Structure of CahJ in Complex with Salicyl Adenylate | | Descriptor: | 9-(5-O-{(S)-hydroxy[(2-hydroxybenzene-1-carbonyl)oxy]phosphoryl}-alpha-L-lyxofuranosyl)-9H-purin-6-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Sikkema, A.P, Smith, J.L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ.
Chembiochem, 19, 2018
|
|
5WM5
 
 | | Crystal Structure of CahJ in Complex with 5-Methylsalicyl Adenylate | | Descriptor: | 9-(5-O-{(S)-hydroxy[(2-hydroxy-5-methylbenzene-1-carbonyl)oxy]phosphoryl}-alpha-L-lyxofuranosyl)-9H-purin-6-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Sikkema, A.P, Smith, J.L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ.
Chembiochem, 19, 2018
|
|
5WPR
 
 | | Crystal structure HpiC1 in C2 space group | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WPP
 
 | | Crystal structure HpiC1 W73M/K132M | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5W7R
 
 | | Crystal structure of OxaC in complex with SAH and oxaline | | Descriptor: | (3E,7aR,12aS)-3-[(1H-imidazol-4-yl)methylidene]-6,12-dimethoxy-7a-(2-methylbut-3-en-2-yl)-7a,12-dihydro-1H,5H-imidazo[1 ',2':1,2]pyrido[2,3-b]indole-2,5(3H)-dione, OxaC, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | Deposit date: | 2017-06-20 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
5WGY
 
 | | Crystal Structure of MalA' C112S/C128S, malbrancheamide B complex | | Descriptor: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
