1CI3
 
 | | CYTOCHROME F FROM THE B6F COMPLEX OF PHORMIDIUM LAMINOSUM | | Descriptor: | HEME C, PROTEIN (CYTOCHROME F), ZINC ION | | Authors: | Carrell, C.J, Schlarb, B.G, Howe, C.J, Bendall, D.S, Cramer, W.A, Smith, J.L. | | Deposit date: | 1999-04-07 | | Release date: | 1999-08-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the soluble domain of cytochrome f from the cyanobacterium Phormidium laminosum.
Biochemistry, 38, 1999
|
|
3BOF
 
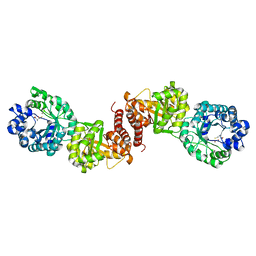 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima complexed with Zn2+ and Homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, POTASSIUM ION, ... | | Authors: | Koutmos, M, Smith, J.L, Ludwig, M.L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1CTM
 
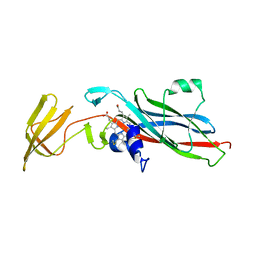 | | CRYSTAL STRUCTURE OF CHLOROPLAST CYTOCHROME F REVEALS A NOVEL CYTOCHROME FOLD AND UNEXPECTED HEME LIGATION | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Martinez, S.E, Huang, D, Szczepaniak, A, Cramer, W.A, Smith, J.L. | | Deposit date: | 1994-01-02 | | Release date: | 1994-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation.
Structure, 2, 1994
|
|
1JVN
 
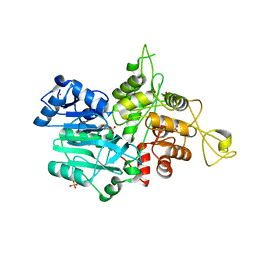 | | CRYSTAL STRUCTURE OF IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE: A TUNNEL THROUGH A (BETA/ALPHA)8 BARREL JOINS TWO ACTIVE SITES | | Descriptor: | BIFUNCTIONAL HISTIDINE BIOSYNTHESIS PROTEIN HISHF, NICKEL (II) ION, PYROPHOSPHATE 2-, ... | | Authors: | Chaudhuri, B.N, Smith, J.L, Davisson, V.J, Myers, R.S, Lange, S.C, Chittur, S.V. | | Deposit date: | 2001-08-30 | | Release date: | 2001-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of imidazole glycerol phosphate synthase: a tunnel through a (beta/alpha)8 barrel joins two active sites.
Structure, 9, 2001
|
|
3F5H
 
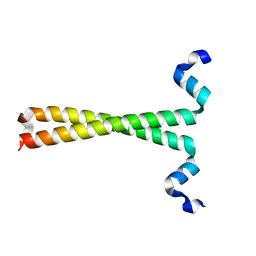 | | Crystal structure of fused docking domains from PikAIII and PikAIV of the pikromycin polyketide synthase | | Descriptor: | SODIUM ION, Type I polyketide synthase PikAIII, Type I polyketide synthase PikAIV fusion protein | | Authors: | Buchholz, T.J, Geders, T.W, Bartley, F.E, Reynolds, K.A, Smith, J.L, Sherman, D.H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for binding specificity between subclasses of modular polyketide synthase docking domains.
Acs Chem.Biol., 4, 2009
|
|
6B3A
 
 | |
5THY
 
 | |
6B39
 
 | | AprA Methyltransferase 1 - GNAT in complex with SAH | | Descriptor: | AprA Methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Skiba, M.A, Smith, J.L. | | Deposit date: | 2017-09-21 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | A Mononuclear Iron-Dependent Methyltransferase Catalyzes Initial Steps in Assembly of the Apratoxin A Polyketide Starter Unit.
ACS Chem. Biol., 12, 2017
|
|
6B3B
 
 | | AprA Methyltransferase 1 - GNAT in complex with Mn2+ , SAM, and Malonate | | Descriptor: | AprA Methyltransferase 1, GLYCEROL, MALONATE ION, ... | | Authors: | Skiba, M.A, Smith, J.L. | | Deposit date: | 2017-09-21 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Mononuclear Iron-Dependent Methyltransferase Catalyzes Initial Steps in Assembly of the Apratoxin A Polyketide Starter Unit.
ACS Chem. Biol., 12, 2017
|
|
5TZ7
 
 | |
5TZ6
 
 | |
5TZ5
 
 | |
1RHY
 
 | | Crystal structure of Imidazole Glycerol Phosphate Dehydratase | | Descriptor: | ACETIC ACID, ETHYL MERCURY ION, GLYCEROL, ... | | Authors: | Sinha, S.C, Chaudhuri, B.N, Burgner, J.W, Yakovleva, G, Davisson, V.J, Smith, J.L. | | Deposit date: | 2003-11-14 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of imidazole glycerol-phosphate dehydratase: duplication of an unusual fold
J.Biol.Chem., 279, 2004
|
|
6D5X
 
 | | Structure of Human ATP:Cobalamin Adenosyltransferase bound to ATP, Adenosylcobalamin, and Triphosphate | | Descriptor: | 5'-DEOXYADENOSINE, ADENOSINE-5'-TRIPHOSPHATE, COBALAMIN, ... | | Authors: | Dodge, G.J, Campanello, G, Smith, J.L, Banerjee, R. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sacrificial Cobalt-Carbon Bond Homolysis in Coenzyme B12as a Cofactor Conservation Strategy.
J. Am. Chem. Soc., 140, 2018
|
|
6D6Y
 
 | |
5VE4
 
 | | Crystal structure of persulfide dioxygenase-rhodanese fusion protein with rhodanese domain inactivating mutation (C314S) from Burkholderia phytofirmans | | Descriptor: | BpPRF, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Motl, N, Skiba, M.A, Smith, J.L, Banerjee, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation.
J. Biol. Chem., 292, 2017
|
|
5VE5
 
 | | Crystal structure of persulfide dioxygenase rhodanese fusion protein with rhodanese domain inactivating mutation (C314S) from Burkholderia phytofirmans in complex with glutathione | | Descriptor: | BpPRF, CHLORIDE ION, FE (III) ION, ... | | Authors: | Motl, N, Skiba, M.A, Smith, J.L, Banerjee, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation.
J. Biol. Chem., 292, 2017
|
|
6D5K
 
 | | Structure of Human ATP:Cobalamin Adenosyltransferase bound to ATP, and Adenosylcobalamin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-DEOXYADENOSINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dodge, G.J, Campanello, G, Smith, J.L, Banerjee, R. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Sacrificial Cobalt-Carbon Bond Homolysis in Coenzyme B12as a Cofactor Conservation Strategy.
J. Am. Chem. Soc., 140, 2018
|
|
5THZ
 
 | |
1S48
 
 | | Crystal structure of RNA-dependent RNA polymerase construct 1 (residues 71-679) from BVDV | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Choi, K.H, Groarke, J.M, Young, D.C, Kuhn, R.J, Smith, J.L, Pevear, D.C, Rossmann, M.G. | | Deposit date: | 2004-01-15 | | Release date: | 2004-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1S4F
 
 | | Crystal Structure of RNA-dependent RNA polymerase construct 2 from bovine viral diarrhea virus (BVDV) | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Choi, K.H, Groarke, J.M, Young, D.C, Kuhn, R.J, Smith, J.L, Pevear, D.C, Rossmann, M.G. | | Deposit date: | 2004-01-16 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1S49
 
 | | Crystal Structure of RNA-dependent RNA polymerase construct 1 (residues 71-679) from bovine viral diarrhea virus complexed with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA-dependent RNA polymerase | | Authors: | Choi, K.H, Groarke, J.M, Young, D.C, Kuhn, R.J, Smith, J.L, Pevear, D.C, Rossmann, M.G. | | Deposit date: | 2004-01-15 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5W3V
 
 | | Crystal Structure of macaque APOBEC3H in complex with RNA | | Descriptor: | Apobec3H, RNA (5'-R(P*AP*AP*CP*CP*CP*CP*GP*GP*GP*C)-3'), RNA (5'-R(P*AP*AP*CP*CP*CP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Bohn, J.A, Thummar, K, York, A, Raymond, A, Brown, W.C, Bieniasz, P.D, Hatziioannou, T, Smith, J.L. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA.
Nat Commun, 8, 2017
|
|
5VE3
 
 | | Crystal structure of wild-type persulfide dioxygenase-rhodanese fusion protein from Burkholderia phytofirmans | | Descriptor: | BpPRF, FE (III) ION | | Authors: | Motl, N, Skiba, M.A, Smith, J.L, Banerjee, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation.
J. Biol. Chem., 292, 2017
|
|
5W7P
 
 | | Crystal structure of OxaC | | Descriptor: | OxaC, S-ADENOSYLMETHIONINE | | Authors: | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
